An experimental extremely high-accuracy, ultra-long sequencing kit

We are pleased to announce a new experimental dataset comprising extremely high accuracy, ultra-long sequencing reads, shared during our Nanopore Community Meeting technical update.
The following cell line samples were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research: GM24385
Data location
As with previous releases the new dataset is available for anonymous download from an Amazon Web Services S3 bucket. The bucket is part of the Open Data on AWS project enabling sharing and analysis of a wide range of data.
The data is located in the bucket at:
s3://ont-open-data/gm24385_2023.12/
See the tutorials page for information on downloading the dataset.
Sample preparation and analysis
Ultra-long libraries of native DNA from GM24385 (HG002) were prepared using a modified Ultra-Long DNA Sequencing Kit V14 motor protein and experimental high-accuracy run conditions. Sequencing was performed on a PromethION instrument to obtain 125 Gbp of sequencing data passing quality filters (read Q-score > Q10, see here), with a read length N50 of 91 kbp. This data was basecalled using a bespoke dorado model to yield a median accuracy of Q26.4.
The per-base quality scores produced by the experimental basecaller model have not been calibrated and may not reflect the empirical accuracy of the called bases.
Read alignment accuracy was measured against
the telomere-to-telomere consortium’s HG002 reference sequence using the
bamstats tool from the fastcat package. The histogram outputs
of bamstats was used to produce the plots in Fig. 1.
Genome assembly
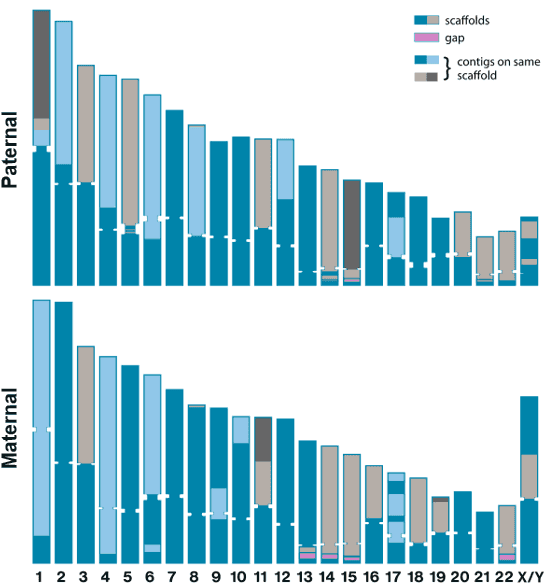
With this dataset, parental sequencing reads, and the Hifiasm and RAFT tools, a diploid human genome assembly was contructed. The assembly included 19 telomere-to-telomere chromosomes. We hope release of this dataset will spurn innovation from assembly algorithm developers as well as serve as a resource for researchers studying the most complicated and inaccessible reaches of the human genome.
| Size / Gbp | Scaffold N50 / Mbp | |
|---|---|---|
| Maternal | 3.03 | 135 |
| Paternal | 2.94 | 133 |
Further information
For additional information regarding these data please contact support@nanoporetech.com.
Related Posts
Information
