EPI2ME Labs 20.10 Release
We are pleased to introduce a new release of our EPI2ME Labs.
With this update we are now supporting only the Jupyter interface to EPI2ME Labs. This Jupyter support has the consequence that users no longer require a Google account to run or save EPI2ME Labs tutorials and EPI2ME Labs is now available to users regardless of their geographic location. Additional information on our reasons for the move to Jupyter are described in a separate post
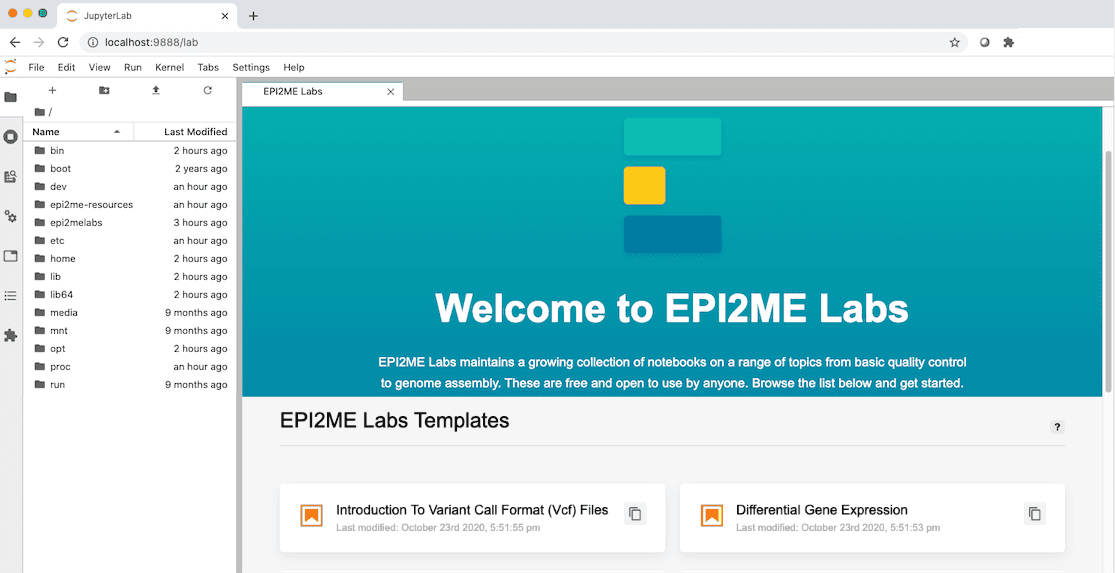
The EPI2ME Labs landing. This provides a link to the available tutorials and workflows that are packaged inside the accompanying docker container. As well as any recent documents of your own
This release includes a number of extensions to the Jupyter software to support usability. These extensions include
Inclusion of autorun code cells that are automatically computed when a notebook is opened. This functionality allows for the preparation of forms for data-entry and for the setting of contextual notebook variables.
The code cell collapser enables larger blocks of computer code to be hidden (or unhidden) within a workflow. This makes the more technical notebooks simpler to read by encouraging focus to results and the written texts.
The play button extension replicates the functionality of the Google Colaboratory button; a play icon is placed on the code cell that is to be run. This is simpler than the default Jupyter play button that may execute code that is elsewhere and outside of the visible screen.
We hope that these extensions may also be of use in the preparation of your own notebooks and workflows. Future EPI2ME Labs releases will continue to further extend the Jupyter environment to simplify bioinformatics analyses and the presentation of results.
All of our EPI2ME Labs tutorials have been converted to the Jupyter flavour. See the Quick start guide guide for instructions of installing and launching the EPI2ME Labs software.
We look forwards to your feedback on this EPI2ME Labs update and we would welcome any recommendations for workflows that you would like to see in upcoming releases.
Tags
Share
Related Posts
Information

