EPI2ME Desktop
Nanopore sequencing can provide a wealth of data. The traditional model of biological data analysis has wet lab scientists producing data and bioinformaticians analysing that data to produce scientific insights. EPI2ME products break this paradigm by enabling data producers to inspect and analyse their data; freeing the time of bioinformaticians for other tasks.The EPI2ME Desktop application is easy to install and allows users of any skill level to run many different workflows.We have workflows ranging from plasmid reconstruction and verification, transcriptomics, through to whole genome variant calling. We have a bit of something for everyone! The full list can be found here.The application runs on your device, where you have full control of your data. If you are lucky enough to have access to a compute cluster, EPI2ME Desktop can even make use of that for those bigger analyses that you might need to run.Users wanting to run large analyses who don't have access to compute clusters can use our hosted compute in EPI2ME cloud. EPI2ME cloud users gain access to their own isolated environment independent of all other users. EPI2ME Desktop integrates seemlessly with EPI2ME cloud. Users can run all the same workflows in their cloud environment as on their local resources.
Data analysis for everyone
- Intuitive interface, seamless experience
- Run preconfigured workflows for rapid insights
- No bioinformatics experience required
- Run in the cloud or locally, on your laptop or preferred machine set-up
- Choose between real-time or post-run analysis
- Keep ownership of your data from sample to answer

An open analysis platform
- Standalone desktop application
- Pre-packaged with >10 open-source workflows (Nextflow framework)
- Compatible with Windows, macOS, and Linux
- Installs on laptop, desktop computer, cluster or cloud service.
- Free access from the intuitive interface or the command line

Intuitive workflows and interactive reports
A full suite of data analysis workflows for:
- Human genomics
- Cancer genomics
- Assembly
- Metagenomics
- Single cell and transcriptomics
- Infectious diseases
- Target sequencing
- And much more…
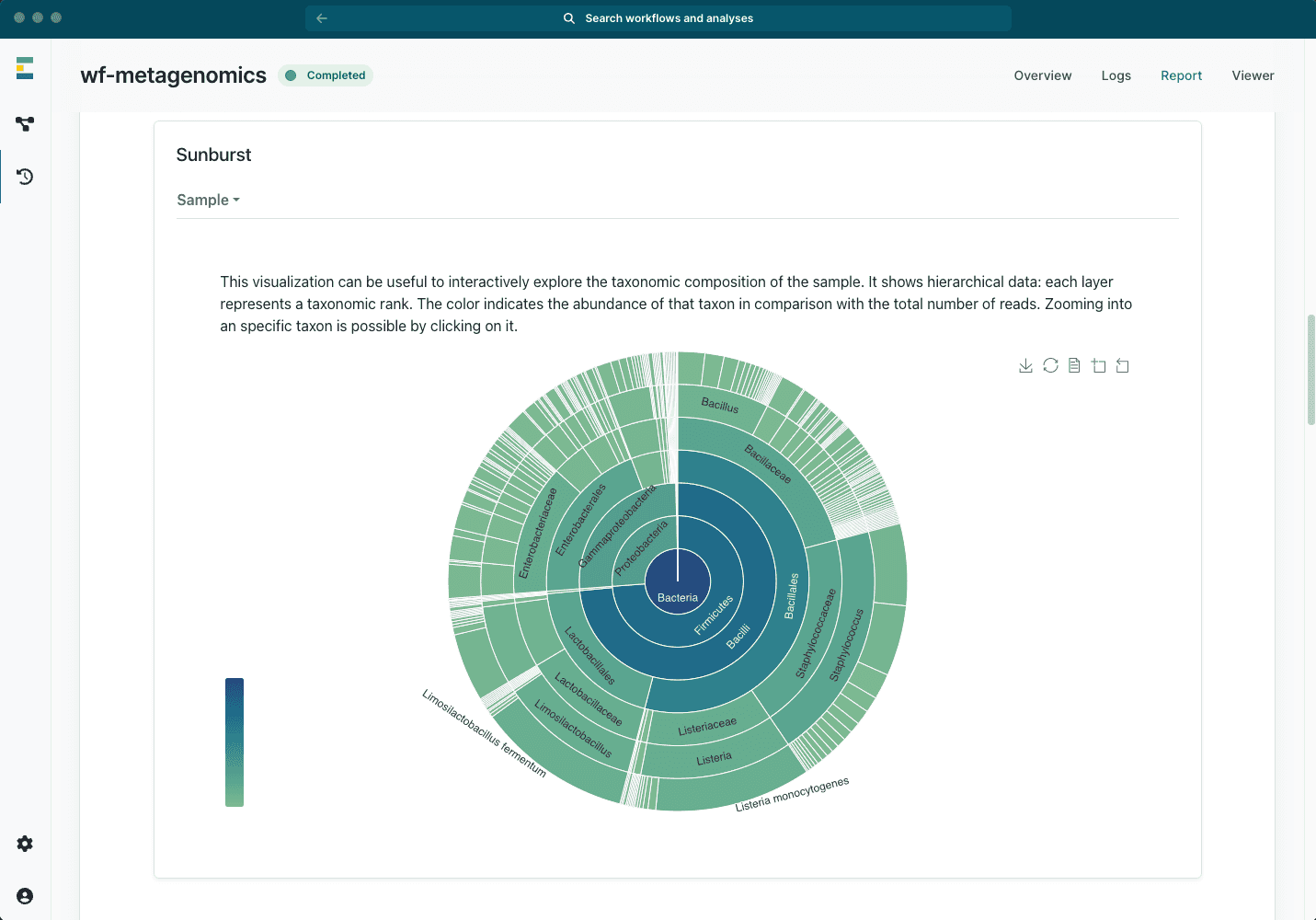
Real-time metagenomics insights
- From amplicon-targeted to shotgun metagenomics sequencing
- Intuitive reports with powerful and interactive graphics
- Real-time results for quick detection
- Post-run analysis for precise identification
- Multiple databases available
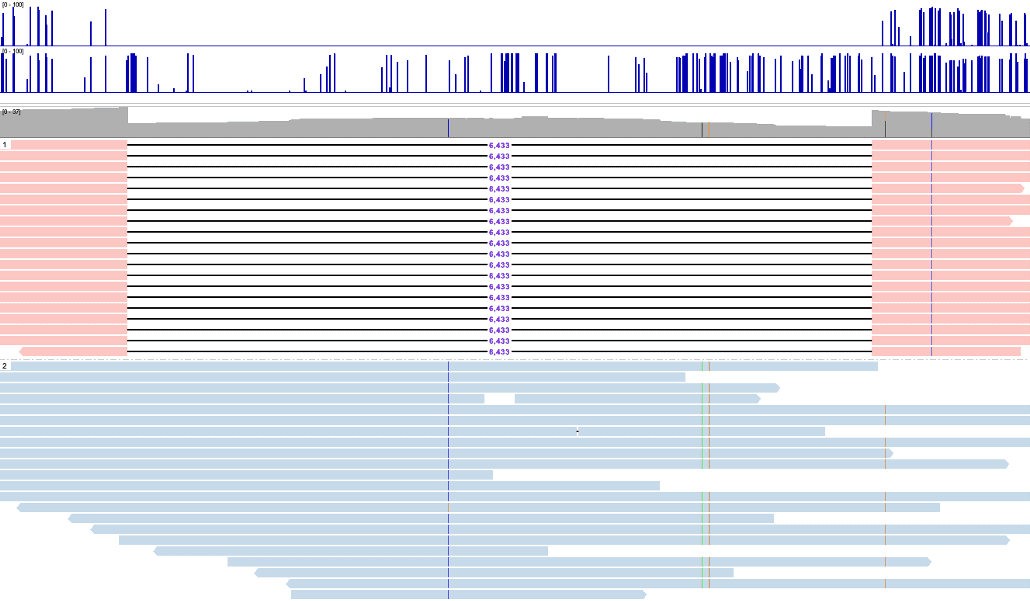
Comprehensive human genome analysis tool
A single workflow to investigate the full human genomic variation
- Single nucleotide variants (SNVs)
- Structural variants (SVs)
- Methylation
- Copy number variants (CNVs)
- Short tandem repeat (STR) expansions
- Phasing
- Comprehensive reports
- Integrated genome browser
© 2020 - 2025 Oxford Nanopore Technologies plc. All rights reserved. Registered Office: Gosling Building, Edmund Halley Road, Oxford Science Park, OX4 4DQ, UK | Registered No. 05386273 | VAT No 336942382. Oxford Nanopore Technologies, the Wheel icon, EPI2ME, Flongle, GridION, Metrichor, MinION, MinIT, MinKNOW, Plongle, PromethION, SmidgION, Ubik and VolTRAX are registered trademarks of Oxford Nanopore Technologies plc in various countries. Oxford Nanopore Technologies products are not intended for use for health assessment or to diagnose, treat, mitigate, cure, or prevent any disease or condition.
