Bed bugs

(Yes, I know that’s not a bed bug; its a coconut rhinoceros beetle.)
The EPI2ME team is happy to release Bed Bugs, an online tool for validating input files for adaptive sampling experiments with MinKNOW. This was a little bit of a fun side project for us using some technologies we’ve previously experimented with but never found a good use for.
Adaptive sampling is a feature unique to Oxford Nanopore Technologies’ sequencing systems. The technique enables a large number of applications to be carried out with very basic sample preparation, leaving the complex target selection to be performed by the sequencer itself. Adaptive sampling can be used to enrich for strands that contain a target region of interest, thereby significantly decreasing the costs while preserving all the benefits of long-read sequencing. Users can also use it to reject strands from an organism which is of no interest. For example in the case of microbiome applications this could provide a simpler workflow, negating any need to deplete the host during sample preparation.
Another use for adaptive sampling is to balance coverage of barcodes, amplicons or regions of a genome ensuring target depths are achieved uniformly for the regions of interest.
Introducing Bed Bugs
Central to the implementation of adaptive sampling within the MinKNOW instrument software is that users must provide reference sequences and a so-called BED file denoting regions of interest. Currently MinKNOW provides little up-front validation of these inputs, which can leave users confused when their adaptive sampling experiment hasn’t achieved what they set out to do. BED Bugs aims to fill this gap by providing some simple formatting checks and validations on these files including cross-checks between the files. It provides the user with feedback as to potential issues with their files.
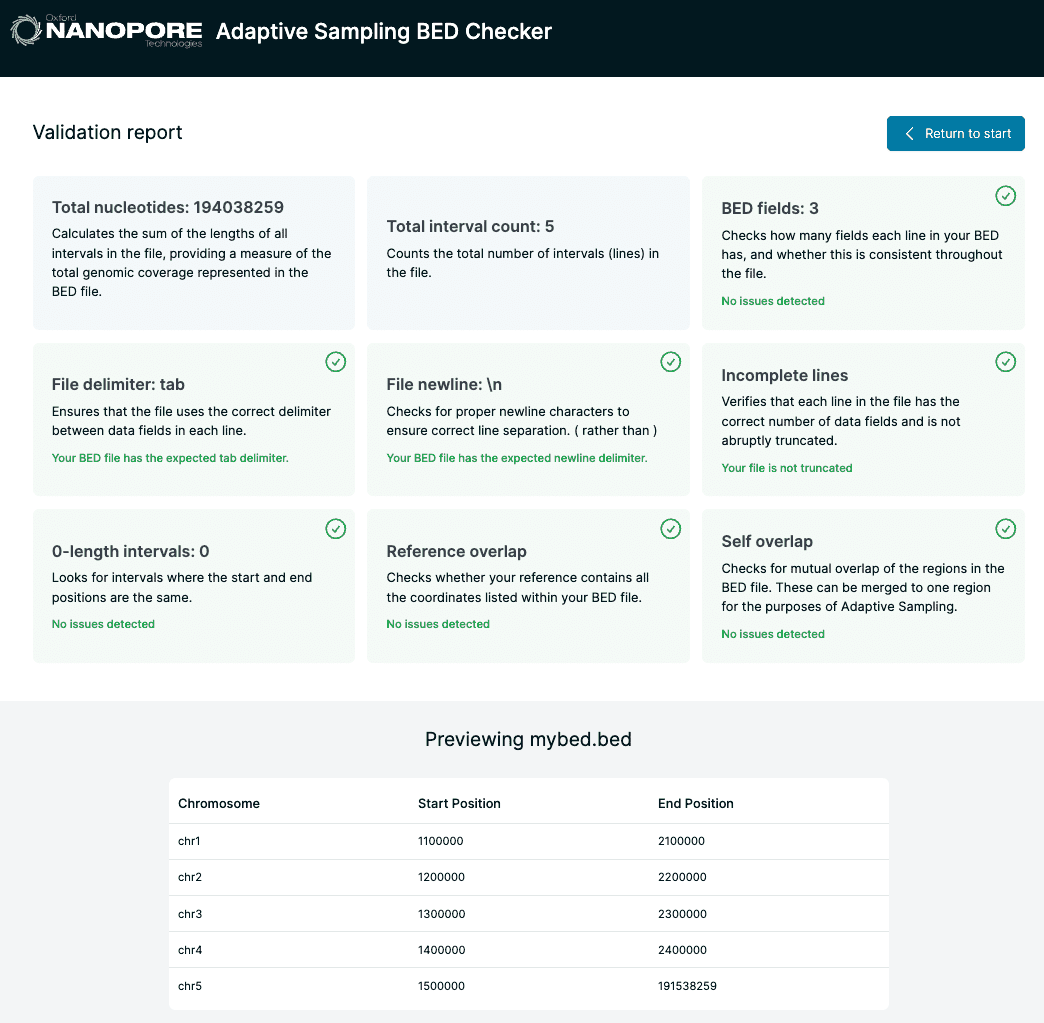
Implementation
For readers who just want to check their adaptive sampling inputs, read no further. You can start using Bed Bugs now!
The core technology used within Bed Bugs is WebAssembly, Wasm for short. Wasm is a binary instruction format designed as a portable compilation target for programming languages. It allows code written in familiar languages such as C to be run in web browsers. WebAssembly aims to execute code at native speed by taking advantage of hardware capabilities of modern processors. By running in a web browser, Wasm allows developers to reach a large audience with their code without worrying about different operating systems and processor types.
Bed Bugs contains WebAssembly components created from bioinformatics codebases
written in C and C++, along with more standard Javascript code,
As BED files are at their heart simply tab-delimited text files, Bed Bugs
uses the robust and popular text-file parser PapaParse
to perform basic checks on the literal file contents.
In order to check the format of the user provided reference files we use
C code including the venerable kseq.h header to create a WebAssembly .fasta
parser.
By successfully parsing the users reference with this library we can assert that
the file is valid.
For the case of the user providing their reference as a minimap2 index (.mmi)
we have a WebAssembly module compiled from custom C code that reads the header
sections of the file.
Both the .fasta and .mmi parsers extract the names and lengths of sequences stored within the file.
Having performed basic checks on the BED and reference file provided by the user,
BED Bugs applies additional contextual validations using bedtools.
Using Bedtools to parse the BED file provides a secondary test on the file’s basic
formatting as well as checking the file is logically consistent for the purposes
of adaptive sampling.
BED Bugs checks that no two intervals in the file intersect; a circumstance which
although technically not an error in all circumstances could be not what the user
intended.
The tool also checks that all the intervals in the BED file correctly represent
valid regions of of the provided reference sequences, again using bedtools.
This check intersects the user’s BED file with a BED file constructed from the .fasta and .mmi
WebAssembly parsers.
Why not use Wasm more?
As part of our mission to simplify bioinformatics, the EPI2ME team has previously dabbled with Wasm to provide bioinformatics tools to our users. We experimented with providing our bioinformatics tutorials as WebAssembly-based web pages, before the most excellent sandbox.bio project did a better job of this than we ever could. As part of this effort we infact contributed tools to the biowasm project.
Larger scale use of WebAssembly is hampered by one key issue: memory use. The WebAssembly runtime is limited to using 4GB of memory, this is prohibitively small for a variety of bioinformatics tasks such as genome assembly and even alignment of reads to large-ish genomes. Solely for this reason we have put Wasm to the side as being a curiosity, worthy of investigation but not ultimately useful to build bioinformatics tools for end-users.
Summary
Bed Bugs is a tools for validating user provided adaptive sampling input files for the MinKNOW device software. It uses technologies that allow the delivery of bioinformatics tools to users without installation or use of a command-line environment. We hope users find the tools useful whilst functionality is being implemented within MinKNOW itself.

