EPI2ME 23.07-01 Release

Dear Nanopore Community,
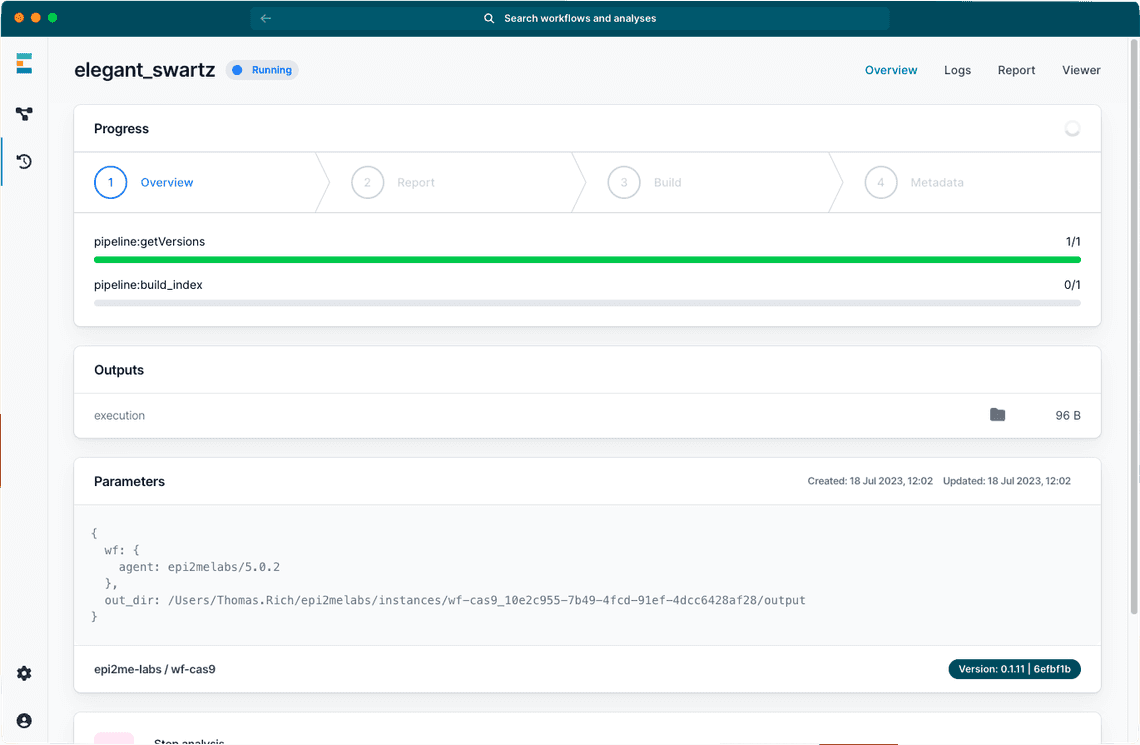
We’re pleased to bring user experience updates to our EPI2ME desktop application (v5.1.0). For our Microsoft Windows users we have overhauled and simplified the installation process and we are now providing an EPI2ME WSL distribution. To improve feedback while your analysis is running, the application now reports progress bars for our workflows (Figure 1).
Other improvements include:
- Enhanced handling of demonstration data when workflows are updated
- Exporting of log information to assist issue reporting
- Automatic checking for available updates to the application
- Enabling more powerful workflow schemas
The EPI2ME software release is available from the downloads page
As a reminder we are currently soliciting early access users for the new cloud analysis functionality in the EPI2ME desktop application that will launch in 2024. You can register your interest here.
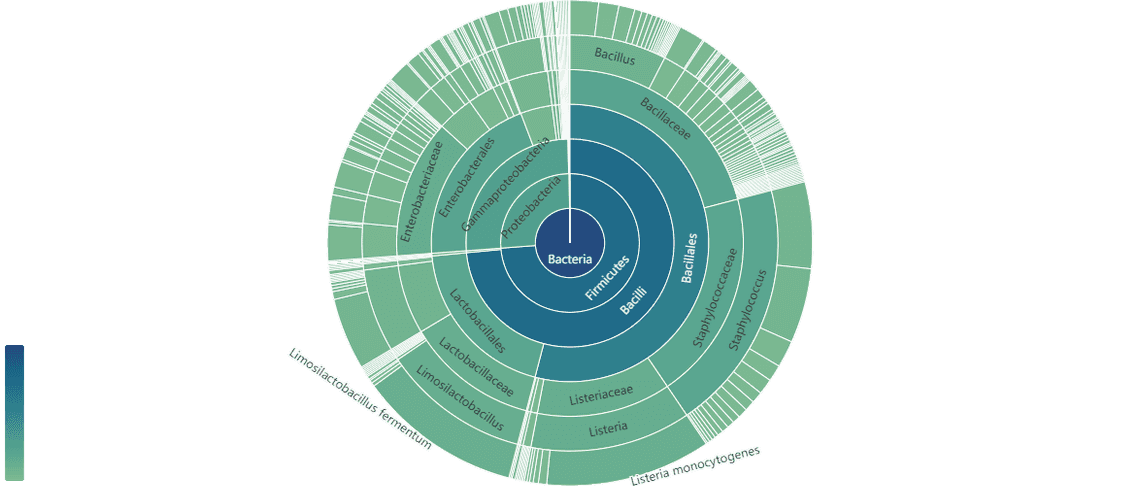
We also have several workflow updates to which we would like to draw your attention. Highlights include wf-metagenomics (v2.4.1) which now provides antimicrobial gene identification, the option to use the SILVA.138 database to classify reads, memory management to solve issues with the PlusPF8 database, and finally, striking sunburst plots to display your data (Figure 2). wf-somatic-variation (v.0.3.0) now includes modified-base basecalling with information on differentially modified loci.
Other workflow updates include:
- wf-clone-validation (v0.4.0) - Now outputs a FASTQ and average quality scores of final assembly and reports any variants between an insert reference and assembly consensus. Also some parameter changes and documentation improvements.
- wf-transcriptomes (v0.2.1) - Workflow has been updated to support only NCBI, Ensembl and Encode genome annotation files for differential gene expression. Also no longer requires a condition sheet.
- wf-cas9 (v0.1.12) - Workflow has been updated to accept input files and directories that include spaces.
- wf-somatic-variation (v0.3.0) - Workflow has been updated to use nanomonsv (v.0.6.0) and now implements the call of modified bases and differentially modified loci and regions with (
--mod.) - wf-metagenomics (v2.4.1) - Workflow has been updated to identify antimicrobial resistance genes using Abricate (through the flag
--amrand--amr_dbto choose among different databases). The workflow now supports memory-mapping large database files so these can be used on devices where the database size exceeds the available system memory. The well-known SILVA database is also available as an alternative database in both pipelines. Further, sunburst plots to explore taxonomic assignments have also been included in the report for the first time. - wf-single-cell (v0.2.6) - Workflow has been updated to fix the incorrect orientations of alignments when processing data from 10x Genomics 5-prime preparation methods.
We look forward to any feedback and welcome recommendations for workflows and EPI2ME features that could be included in future releases. See you in Singapore!
Related Posts
Information

