EPI2ME Labs 22.01.01 Release
Dear Nanopore Community,
This Wednesday we have a trio of announcements; these include a new Nextflow pipeline for gene isoform characterisation, updates to our wf-artic software and an updated dataset release that includes Remora-called 5mC information from GM24385.
New workflow for gene isoform characterisation from transcriptomic sequencing data
We are delighted to introduce a new EPI2ME Labs workflow, wf-isoforms. This workflow provides a robust pipeline for the characterisation of gene isoforms from transcriptomic sequence collections. The workflow is based on, and now supersedes, the pipeline-nanopore-ref-isoforms and pipeline-nanopore-denovo-isoforms.
wf-isoforms can accommodate single or multiplexed sequence collections and provides a simplified and scalable product for the analysis of gene isoforms. The workflow is best run using available genome annotation information (GTF files) to both assign sequenced reads to known gene isoforms and to aid in the discovery of potentially novel isoforms. The workflow can also be run using experimental de novo parameters to assist in the annotation of genes and their isoforms from organisms where little prior genome annotation is available.
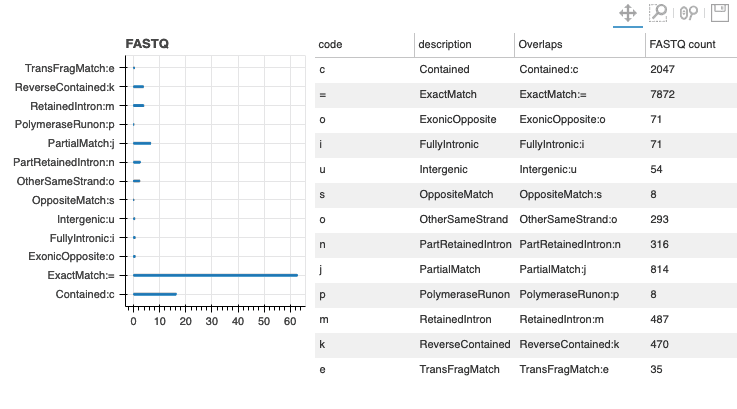
The workflows both use the pychopper software to select for appropriate full-length sequence reads from the starting sequence collection. The workflow produces an HTML format report that summarises the analysis and results obtained. When a reference genome annotation has been used, the results include GffCompare assignments for the observed transcripts; these assignments can be used to identify the potentially novel isoforms as shown in Figure 1.
Updates to wf-artic workflow for SARS-CoV-2 sequence analysis
A new version of
our wf-artic software has also been released. The wf-artic v0.3.10 update
includes support for NEB primersets and includes updates for both Pangolin
(v3.1.17) and Nextclade (v.1.8.0). This update is available through the
project’s github pages and through our EPI2ME product. This release also allows
you to specify –update_data at runtime, which will provide you with the latest
Pangolin and Nextclade tools and datasets. Please also have a review of our blog post
on lineage and clade assignment using wf-artic: SARS-CoV-2 Midnight Analysis.
Remora for 5mC analysis and associated data release
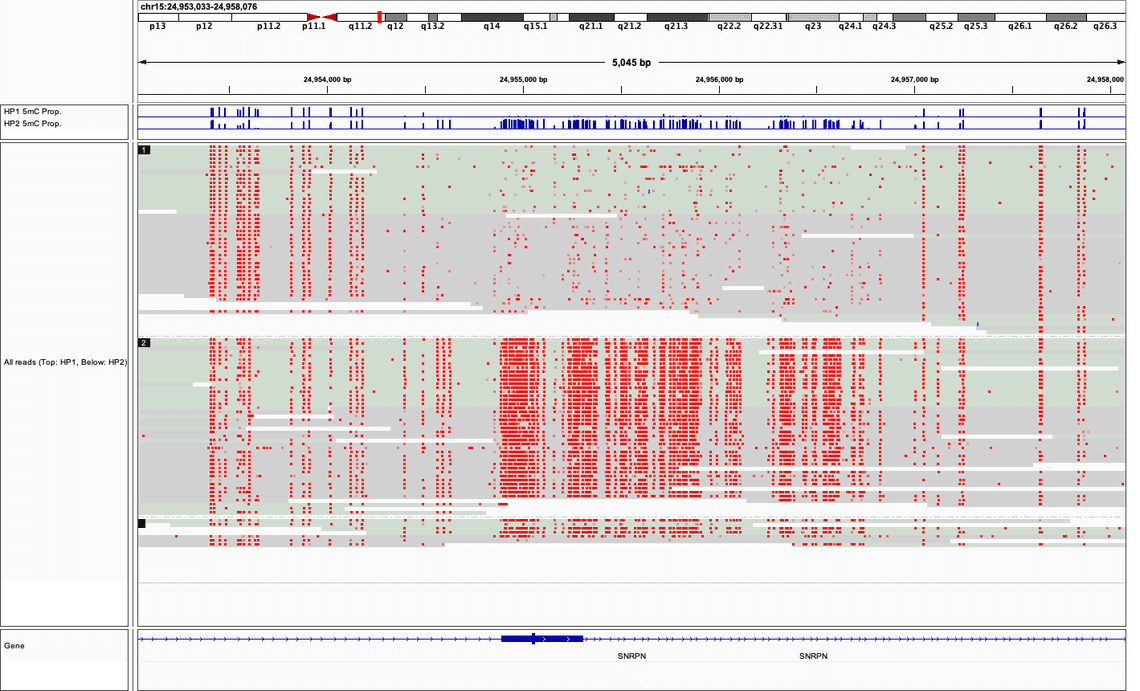
We have also released an ont-open-data dataset that can be used to evaluate and benchmark the 5mC basecalling results obtained using the new Remora algorithm as implemented in Bonito. This dataset and instructions for how it may be used are included in an EPI2ME Labs blog post. The EPI2ME Labs modified bases tutorial has been updated and now uses modbam2bed for the preparation of bedMethyl format data. The tutorial will demonstrate how to produce the beautiful plots as presented in the blog post.
We look forwards to any feedback and comments and would welcome insight as to workflows and tutorials that you would like to see in the future.

