EPI2ME Labs 23.03-01 Release

Our Early March bioinformatics release brings new functionality to the wf-human-variation workflow, our Nextflow pipeline for the analysis of human genetic variation, which has been updated to release v1.3.0. This provides a new module for genotyping short tandem repeat (STR) expansions, a type of genomic variation linked to repeat expansion disorders. The STR genotyping is based on the Straglr software and relies on the phasing information provided within the mapped BAM file to count the repeat units for both maternal and paternal alleles. The HTML reports prepared by the workflow describe the median repeat count for each of the phased alleles and provides additional plots to reveal distribution of repeat lengths across loci tested; this may highlight repeat instability. Please see Figures 1 and 2 for example results that have been observed following an analysis of sequence data from the Coriell NA07063 cell line.
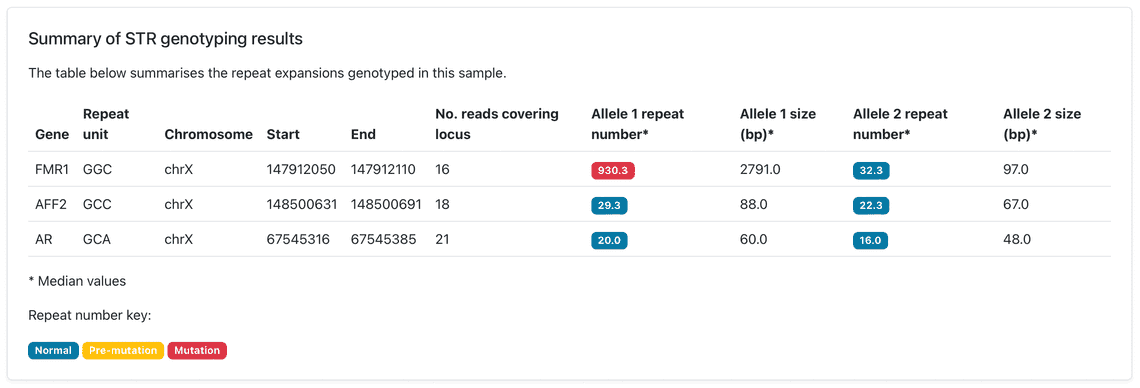
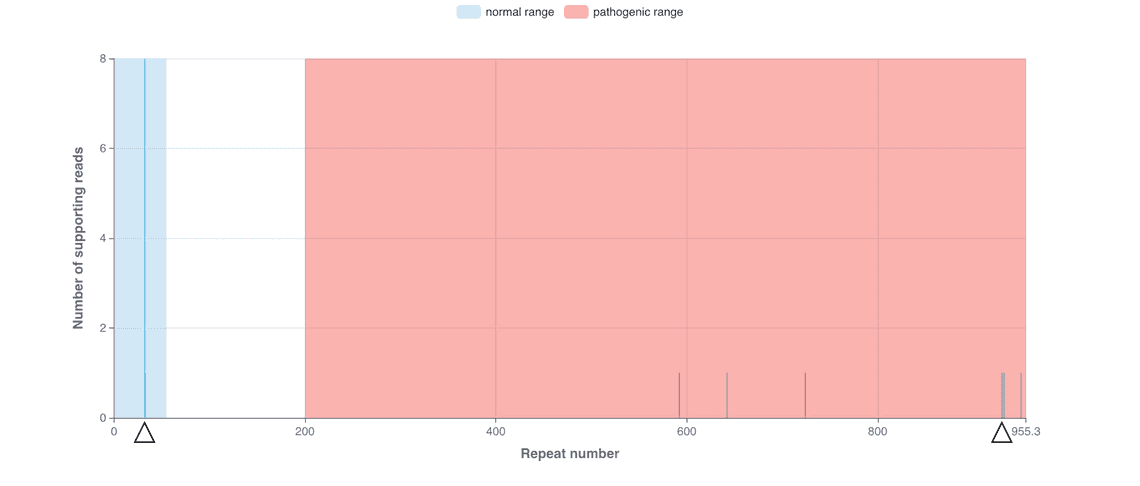
The following cell line samples were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research: NA07063
Other software and workflow updates include:
- The EPI2ME Labs desktop app has been updated to revision v4.1.3. This includes fixes to ensure that the correct version of Java is downloaded on MacOS. Additional checks have been included to prevent more than one EPI2ME Labs instance running at the same time.
- wf-human-variation (v1.3.0)
- Addition of new STR genotyping module.
- wf-cas9 (v0.1.9)
- Improved computer memory usage, especially when processing larger datasets.
- The HTML report has been updated to provide better usability and navigation across datasets.
- wf-single-cell (v0.2.1)
- Performance improvements during analysis steps that include barcode extraction, sequencing saturation calculation, and UMAP generation.
- Bug fix to correct shifted cell barcode and UMI quality scores.
- wf-artic (v0.3.22)
- Pangolin updated to v4.2.
- Nextclade updated to v2.11.0 (new datasets also provided).
- wf-pore-c (v0.0.2)
- Substantial workflow performance updates with updated pore-c-py package (cut run time from 3 days to 2 hours).
- Fix for missing references in pairs file.
- Fixes in simulated paired-end output BAM file.
- wf-metagenomics (v2.0.9)
- Updates to recommended latest kraken2 databases and checks to ensure that appropriate NCBI taxdump is used.
- Provide (original and rarefied, i.e. all the samples have the same number of reads) abundance tables listing taxa per sample for a given taxonomic rank.
- Add alpha diversity indices and richness curves – see Figure 3 for an example.
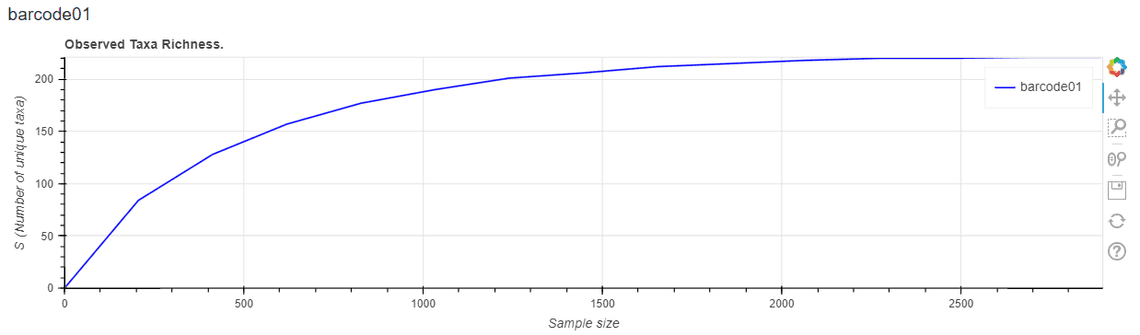
**Figure 3.** Sample-based rarefaction curves have been added to the HTML report to display observed taxa richness. This information may be used to assess whether additional sequencing would substantially influence the observed taxonomic diversity.
- Finally we have released a new bamindex program as part of our fastcat conda package. bamindex creates index files for non-sorted (typically unaligned) BAM files. It may be of use for bioinformatics workflow developers who wish to parallelise operations on such unaligned files. It complements the functionality in the pre-existing bri package.
We would welcome any feedback and would be delighted to receive recommendations for future workflows or datasets.

