EPI2ME 23.05-02 Release

Dear Nanopore Community,
With this post London Calling ‘23 bioinformatics product release, we are delighted to introduce the updated EPI2ME desktop application that was described by Chris Wright during his Data For Breakfast presentation - this is available online.
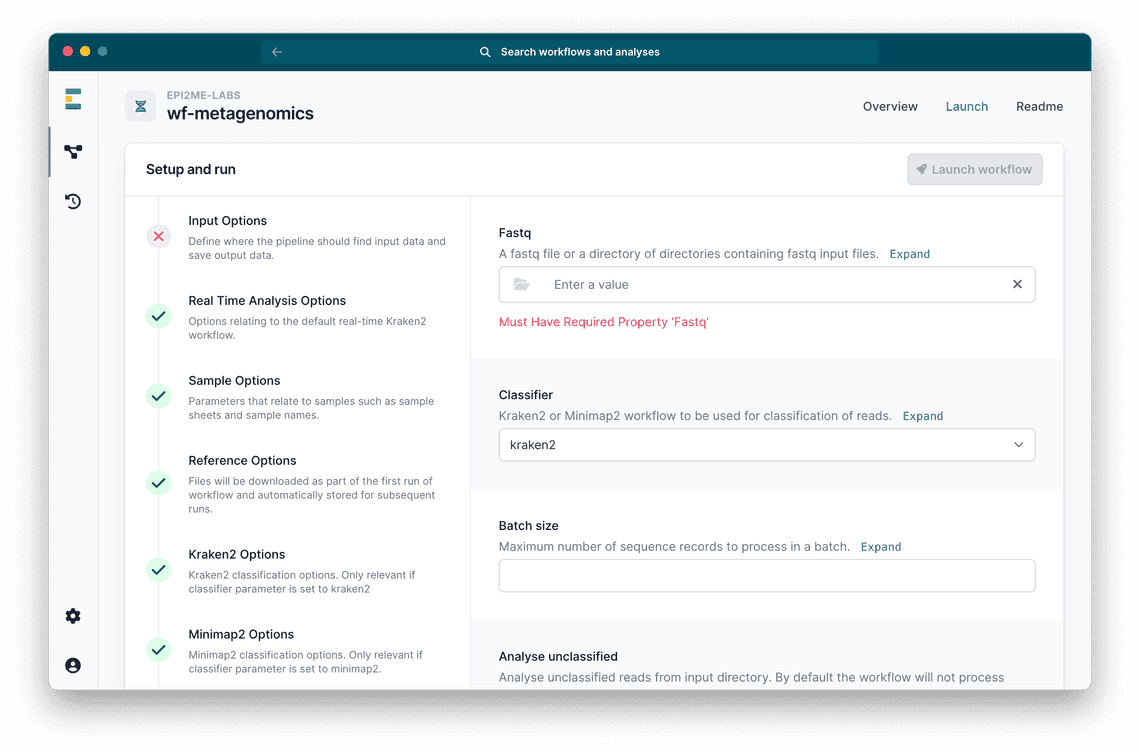
This update provides a simplified user interface to aid the entry of paths and parameters required by workflows (example shown in figure 1). Updates in the future will include new functionality to run analyses in the cloud. Please register your interest for this cloud functionality - we’ll soon be looking for test projects.
We are also pleased to introduce two new datasets that are now available through the s3://ont-open-data resource.
- The Ashkenazi trio and HG001 (CEPH/Utah) have been sequenced from libraries prepared using ligation kit 14 and sequenced using the latest 5khz sampling rate - please see the Sequencing Genome in a Bottle samples blog post.
- The COLO829 tumor/normal pair has been prepared using Ligation Sequencing Kit V14; a description of the data release can be found in the Sequencing A Tumor Normal Pair with Ligation Sequencing Kit V14 post - this dataset is a useful reference dataset to explore our recently introduced wf-somatic-variation workflow.
A couple of workflows have also been updated
- wf-basecalling (v0.6.0) now works in real time - the workflow watches the input path and process raw data files as they become available.
- wf-transcriptomes (v0.1.12) has been updated to accommodate different reference annotation file types - annotations in both GFF3 and GTF format (optionally .gz compressed) are now accepted.
We welcome all feedback and recommendations for new workflows and functionality. Please register your interest and we’ll tell you more about the upcoming cloud functionality.
Related Posts
Information

