EPI2ME Labs Quick Start
This guide pertains to EPI2ME Labs version 3. For a guide to working with the latest version of EPI2ME Labs please see EPI2ME Labs Quick Start.
EPI2ME Labs extends the JupyterLab notebook framework with a pre-configured analysis environment and the ability to run Nextflow workflows all in a single desktop application. Our Nextflow workflows can be used to routine analyses, while our notebooks provide an introduction to nanopore sequencing bioinformatics and provide next steps for a selection of our workflows.
EPI2ME Labs
The first time EPI2ME Labs is started the environment components must be downloaded. This will not happen on subsequent server starts. After the components have been downloaded the Lab environment will start.
Running a Workflow
- On the landing page select the workflow you would like to open.
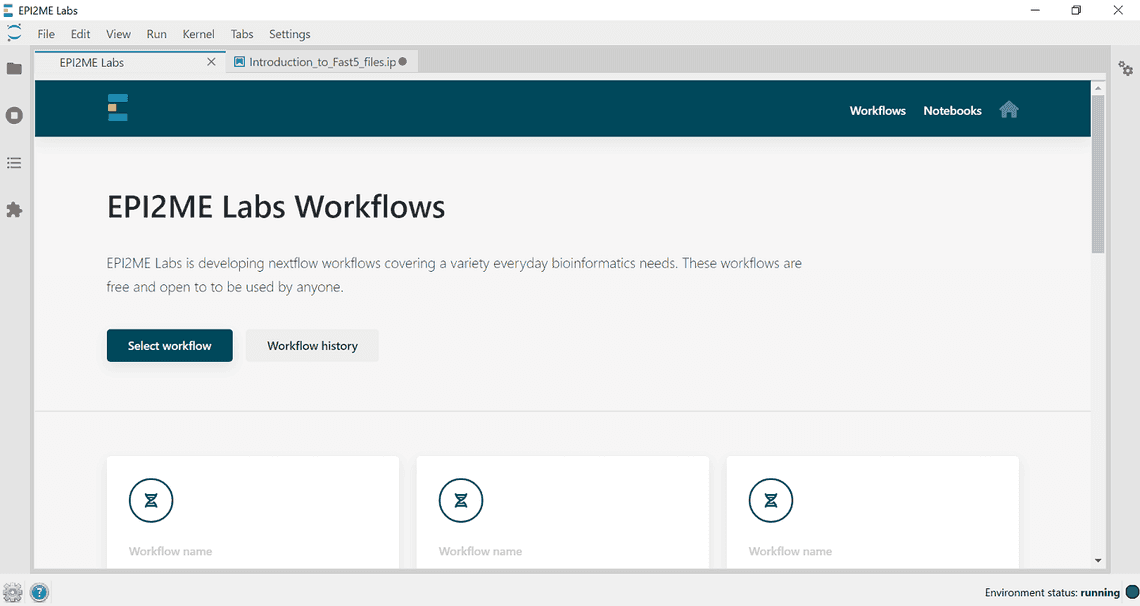
Launch a workflow - Scroll through the form and fill out required fields, required fields will be indicated with a red cross, which will turn to a green tick when filled out correctly. Click Launch workflow.
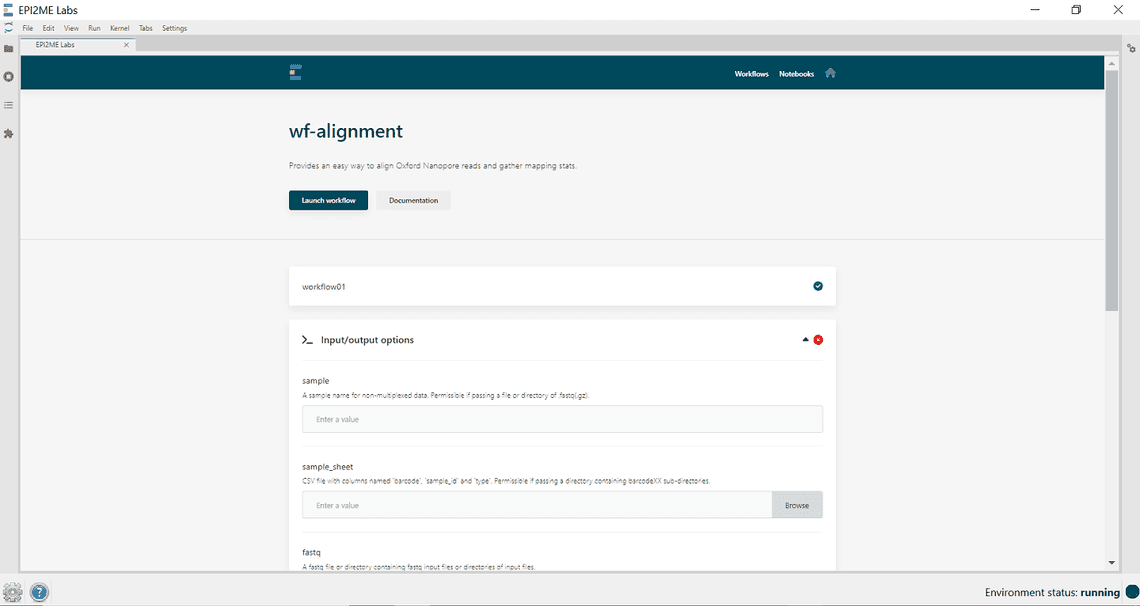
Launch a workflow - The workflow should start running and give an indication it is has launched and when complete it should provide output files.
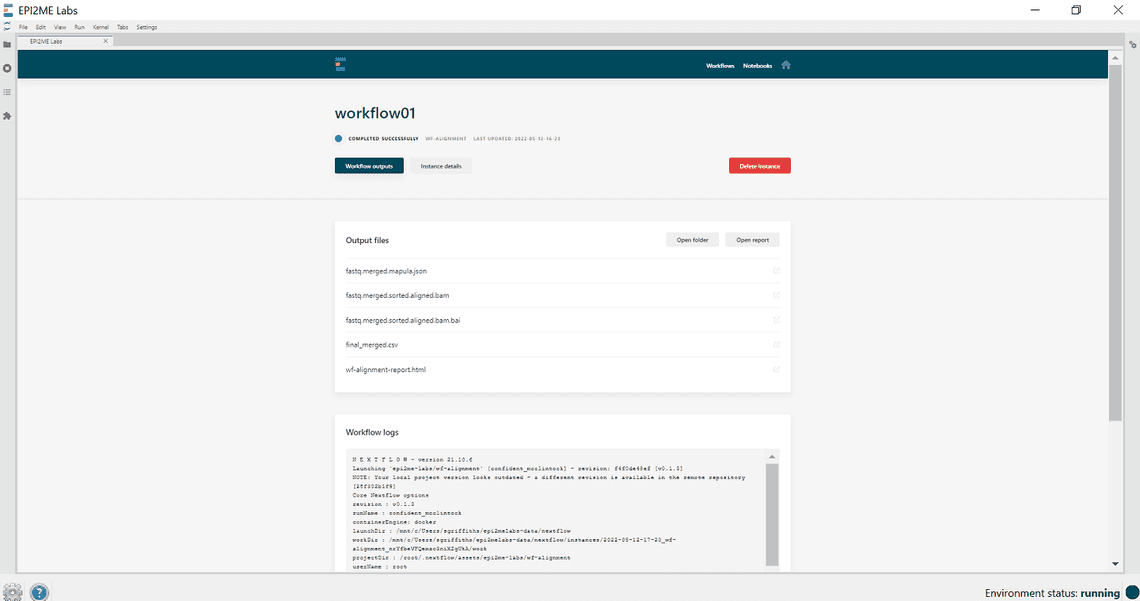
Launch a workflow - Click on instance details for further information about the run and for an option to re-run.
Running a Tutorial
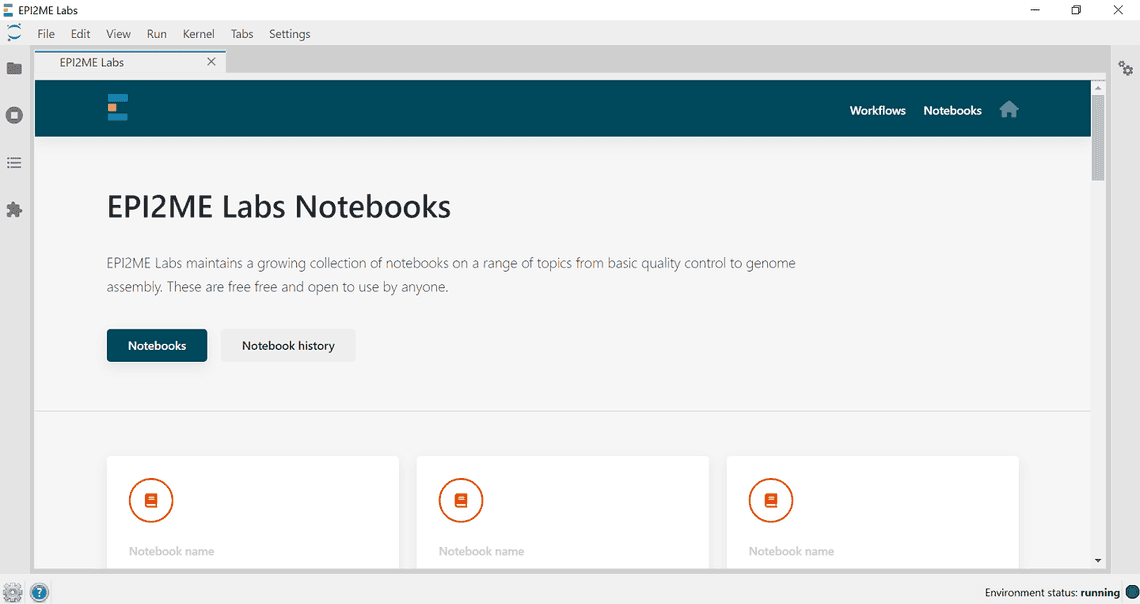
- On the landing page select notebooks in the top right corner and select the notebook template you wish to open. The template will create a copy of the file as a new document for you to edit.
Launch a notebook - You can see all your data in the location you provided in settings by
clicking /epi2melabs in the sidebar:
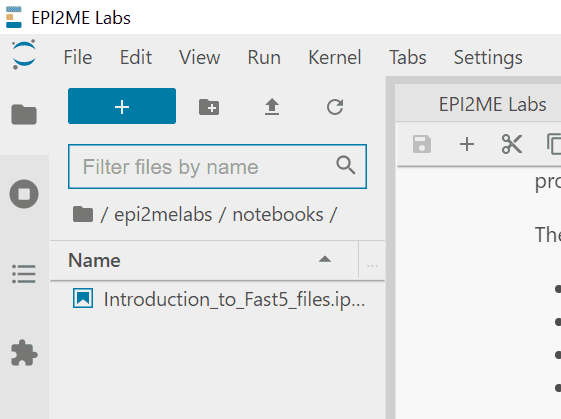
Data access from epi2melabs folder - To copy your own data into the tutorial, right-click on the data file and
select Copy path.
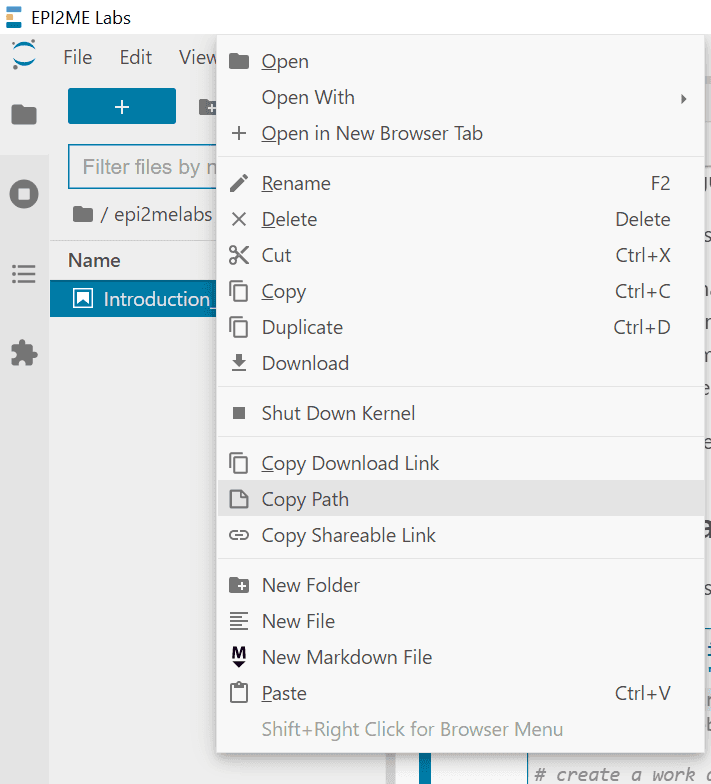
Copying filepaths - Follow the guidance provided in the tutorial to perform data analysis.
Running code cells
Running code cells requires clicking the play button in the navigation bar at the top of the screen (or to the right-hand side of a code cell). There is also a “Run” tab that can be used to run multiple cells.
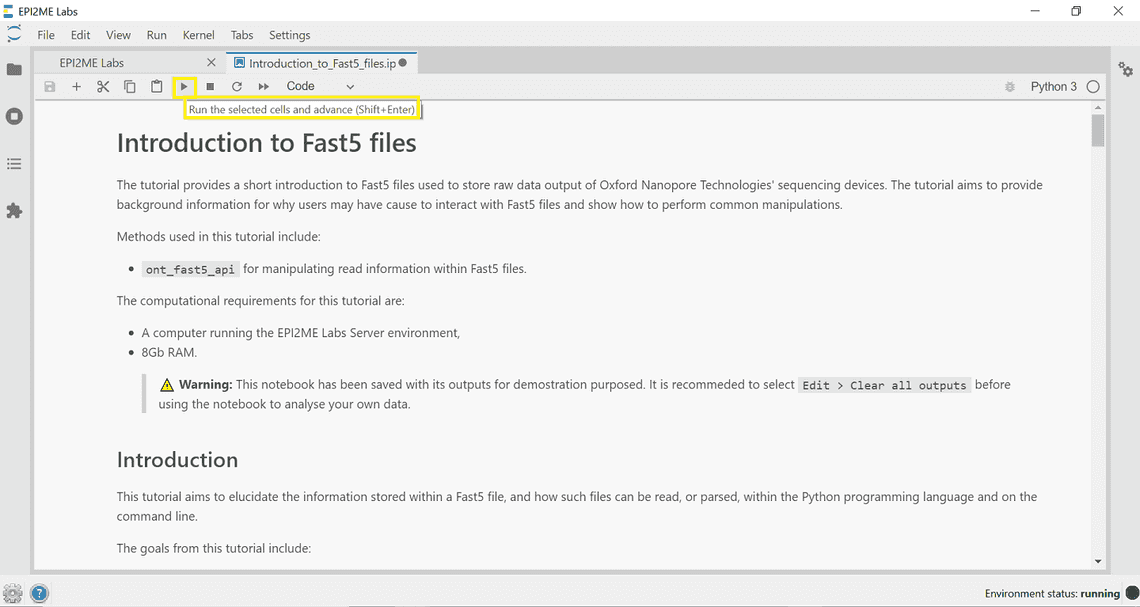
Bookmarks and navigating notebooks
The environment provides a table of contents menu in the left-hand sidebar. These navigation prompts can also be used to fold content visibility in the notebook.
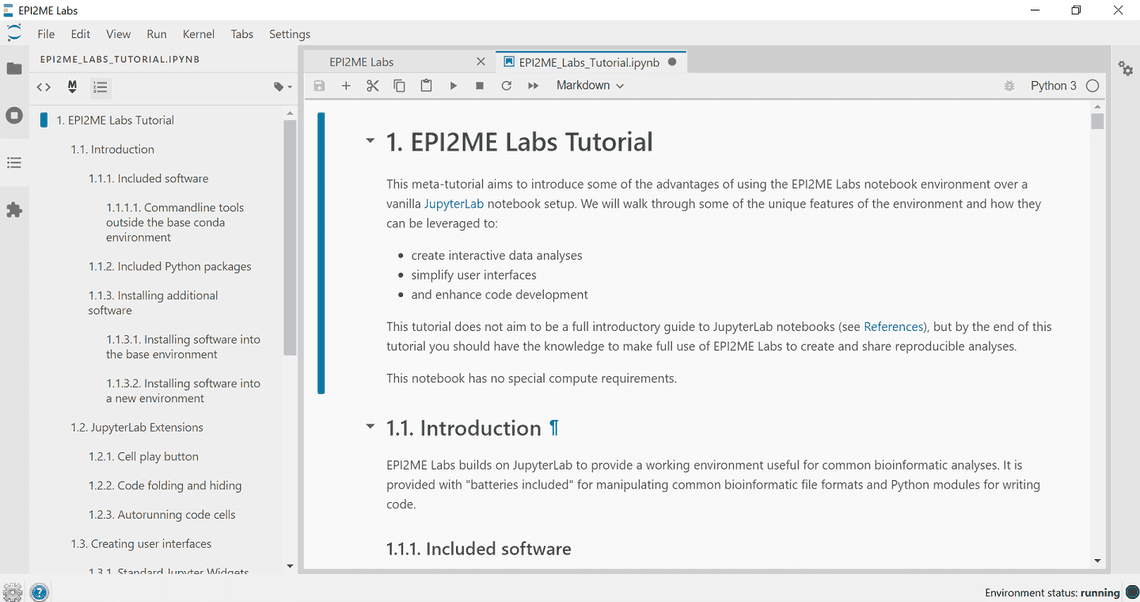
Updating the Environment
The EPI2ME Labs application automatically detects when updates are available to the notebok server. Oxford Nanopore Technologies’ may occassionally provide updates to provide new features or improved performance.
When an update is available attempting to start or restart the notebook server when an update is available will result in the following being displayed:
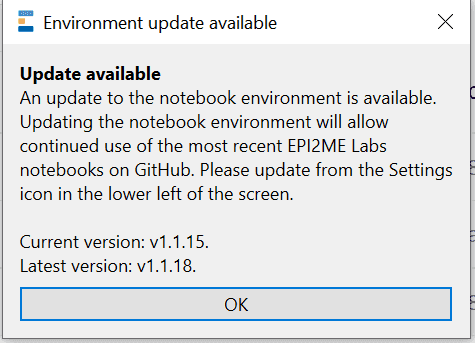
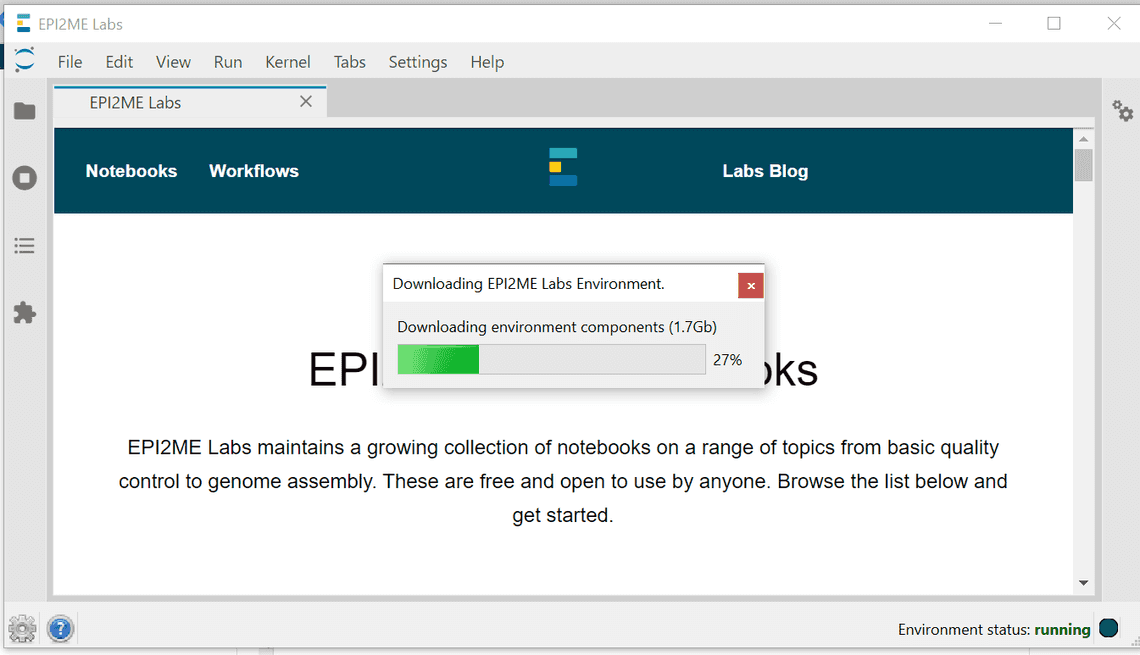
To update the server or launcher select update from the setting menu (bottom left hand cog), The new components will be downloaded:
Application settings
To access the application settings click the bottom left hand cog and click Settings.
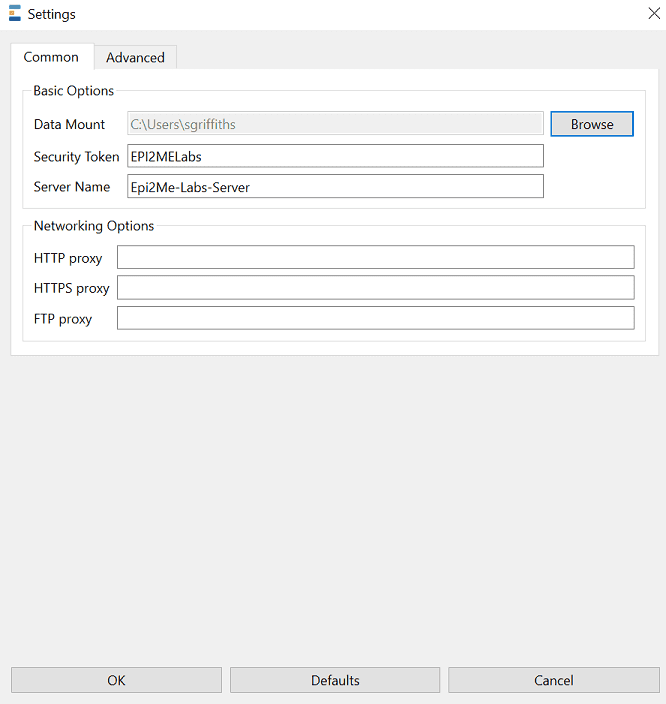
The Data Mount option controls the path where data from Nextflow workflows and Jupyter notebooks is save.
We strongly encourage updating the security token to a unique value. The token provides an authentication method that allows only trusted users access to the EPI2ME Labs server and is used as an additional security layer to help prevent other users on the network from running code and accessing data. This token will be used later in the workflow to connect Google Colaboratory to the server.
Changing these settings will cause the Lab environment to restart.
