Bonito basecalling with R9.4.1
Updated 2020-10-30: This page was edited to reflect the formal release of Bonito v0.3.0
We are please to announce the addition of bonito basecalling results to the GM24385 dataset. Bonito is a research-grade, open source basecaller utilising the PyTorch library; its development explores alternative basecalling frameworks to those use in the product-grade Guppy basecalling software.
The Bonito basecalling for the GM24385 dataset was performed using version
0.3.0, driven by the same katuali analysis pipeline
as for the initial dataset release. The Bonito basecaller was provided as
input the per-chromosome .fast5 files created in the initial pipeline via
alignment of the Guppy 4.0.11 basecalls. This allows for easy comparison of
results on subsets of the data (but may lead to subtle side-effects). For
example the analysis data structure contains now entries of the form:
gm24385_2020.09/analysis/r9.4.1/{flowcell}/guppy_{suffix}/align_unfiltered/{chromosome}/bonito_v0.3.0/├── align_unfiltered│ ├── align_to_ref.log│ ├── basecall_stats.log│ ├── calls2ref.bam│ ├── calls2ref.bam.bai│ └── calls2ref_stats.txt├── basecalls.fastq.gz└── basecalls.fastq.gz_summary.tsv
The file basecalls.fastq.gz contains the basecalling results from Bonito. The
quality scores in these files have been mocked as the pre-release build of Bonito used
does not yet provide quality scores. Similar to the main folder structure the
align_unfiltered directory contains unfiltered alignments of the basecalls to
the reference sequence (calls2ref.bam) along with text files summarizing the
properties of the alignments.
Comparison with Guppy 4.0.11 basecalls
As a basis for comparison with the current Guppy basecaller we can use the alignment summary files for both the Guppy and Bonito basecalls. To simplify the analysis we compare only chromosome 1 data for a single flowcell; we can download the files with:
aws s3 cp --no-sign-request s3://ont-open-data/gm24385_2020.09/analysis/r9.4.1/20200914_1354_6B_PAF27096_e7c9eae6/guppy_v4.0.11_r9.4.1_hac_prom/align_unfiltered/chr1/calls2ref_stats.txt guppy.statsaws s3 cp --no-sign-request s3://ont-open-data/gm24385_2020.09/analysis/r9.4.1/20200914_1354_6B_PAF27096_e7c9eae6/guppy_v4.0.11_r9.4.1_hac_prom/align_unfiltered/chr1/bonito_v0.3.0/align_unfiltered/calls2ref_stats.txt bonito.stats
The following python code,
from concurrent.futures import ProcessPoolExecutorimport pandas as pdimport aplanat.utilfrom aplanat import linesdef read_data(args):caller, filename = argsdf = pd.read_csv(filename, sep='\t')xs, ys = aplanat.util.kernel_density_estimate(df['acc'], step=0.05)df = pd.DataFrame({'accuracy':xs, 'density':ys})df['caller'] = callerreturn dfdata_sets = {'bonito': 'bonito.stats','guppy': 'guppy.stats'}with ProcessPoolExecutor() as executor:dfs = list(executor.map(read_data, data_sets.items()))plot = lines.line([df['accuracy'] for df in dfs],[df['density'] for df in dfs],colors=['red', 'blue'],names=['bonito', 'guppy'],xlim=(85,100),x_axis_label='Alignment accuracy',y_axis_label='Density')plot.legend.location = 'top_left'
can be used to plot a kernel density estimate for the read alignment accuracy:
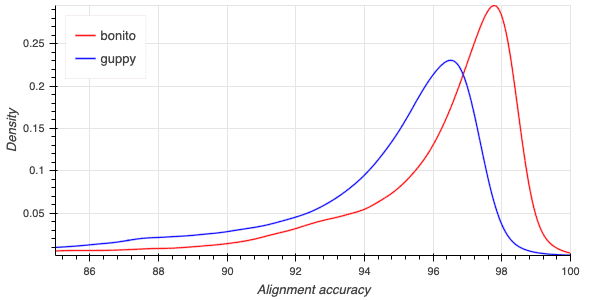
The plot indicates a decrease of one-third in the modal error of reads.

