EPI2ME Labs 22.03.01 Release
Dear Nanopore Community
This week the EPI2ME Labs team is delighted to release a new workflow, wf-bacterial-genomes. We have also released minor updates to wf-artic, wf-alignment and wf-isoforms.
- wf-bacterial-genomes is a Nextflow workflow implemented to simplify bacterial genome assembly. The analysis pipeline performs a quality review on the starting sequence collection prior to assembling the provided sequencing data using Flye. The consensus sequence is polished using Medaka and some basic annotation is performed using Prokka. If a reference genome is provided, the workflow will map sequence reads to this reference using minimap2 and will call variants using medaka variant. This workflow replaces our earlier wf-hap-snp product that performed variant calling in haploid genomes.
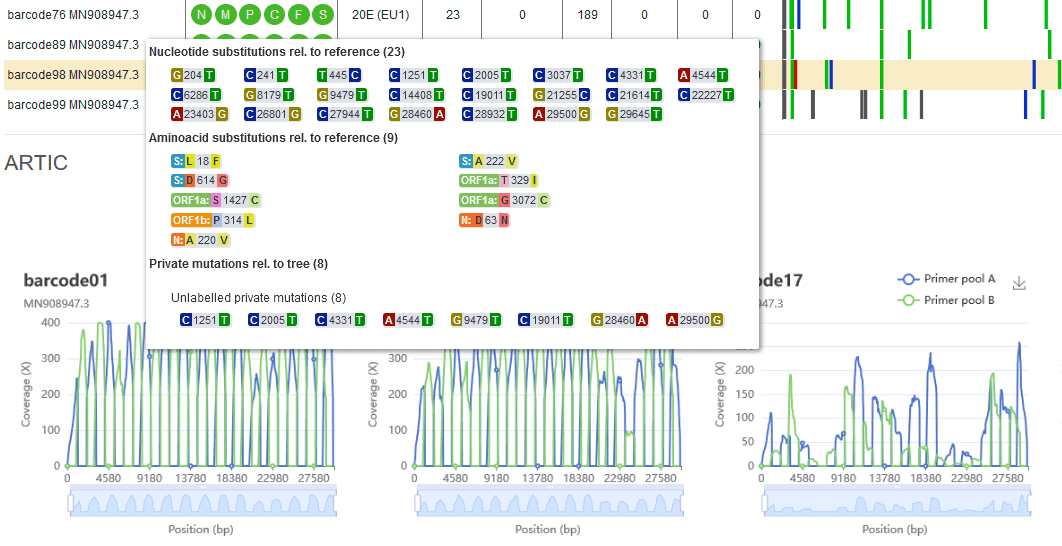
- wf-artic for the analysis of SARS-CoV-2 sequencing data has been updated to version v0.3.12. This release includes an update to the included version of Nextclade (1.10.3) and improvements to the Nextclade visualisation included in the HTML report. Please note that the workflow now uses the r941_min_hac_variant_g507 Medaka model by default. The corresponding EPI2ME workflow, Fastq QC + ARTIC + NextClade, also includes these updates and is available as version v2022.03.08. Please see the product changelog for additional details.
- wf-alignment, our workflow for mapping sequence reads to a reference genome, has been updated to version v0.1.1. This minor update introduces a collection of performance changes - for more details please review the workflow changelog.
- wf-isoforms, for analysis of gene isoforms from cDNA sequencing data, has been updated to version v0.1.1. This update fixes a couple of issues associated with transcript counting and the de novo approach to identifying genes and their isoforms. More information on these updates is provided in the product’s changelog.
- Our metagenomic classification tutorial has been fixed. The bundled Centrifuge software can now find the required indices.
We would welcome any recommendations for future workflows, tutorials and dataset releases.
Related Posts
Information

