EPI2ME Labs Embraces Nextflow
We are excited to launch a new bioinformatics offering using the Nextflow reactive workflow framework. Nextflow has been selected as a preferred framework because of its integration with container technologies, software package managers and its scalability to cluster- and cloud-scale installations. Nextflow also has growing user adoption through projects such as nf-core. These advantages will help us deliver varied workflows with minimal requirements for the installation of additional software.
- wf-artic packages the ARTIC Fieldbioinformatics software in a convenient containerised package that can be used to locally process multiplexed SARS-CoV-2 sequence data in a more automated manner.
- wf-hap-snps is a workflow designed to perform haploid SNP calling from whole genome sequencing of haploid samples.
- wf-alignment packages the minimap2 software and streamlines the process of mapping sequence reads to a reference genome and preparing summary statistics. It can also analyse the abundance of known molarity control experiments and use this information to derive the abundances of other species present in the sample.
- wf-metagenomics includes the Centrifuge software and appropriate indexes to facilitate the taxonomic classification of sequence reads from metagenome samples.
Our Nextflow workflows are intended to build upon the best-practice workflows demonstrated in our EPI2ME Labs tutorials. The workflows use the same methods demonstrated in the tutorials but should be more appropriate and scalable for larger and/or multiplexed datasets. The Nextflow workflows are distributed via GitHub and we hope that users will fork and customise the repositories.
The Nextflow workflows all prepare reports that include summary statistics, tabular data and figures to describe the sequences processed and to highlight important features of the results obtained. Example reports for each of the workflows are available from the Notebook Index.
We support the installation and usage of these workflows on our GridION sequencing devices; the workflows have also been tested on a variety of Linux platforms.
We look forwards to feedback and would welcome requests for workflows and tutorials to be included in future updates.
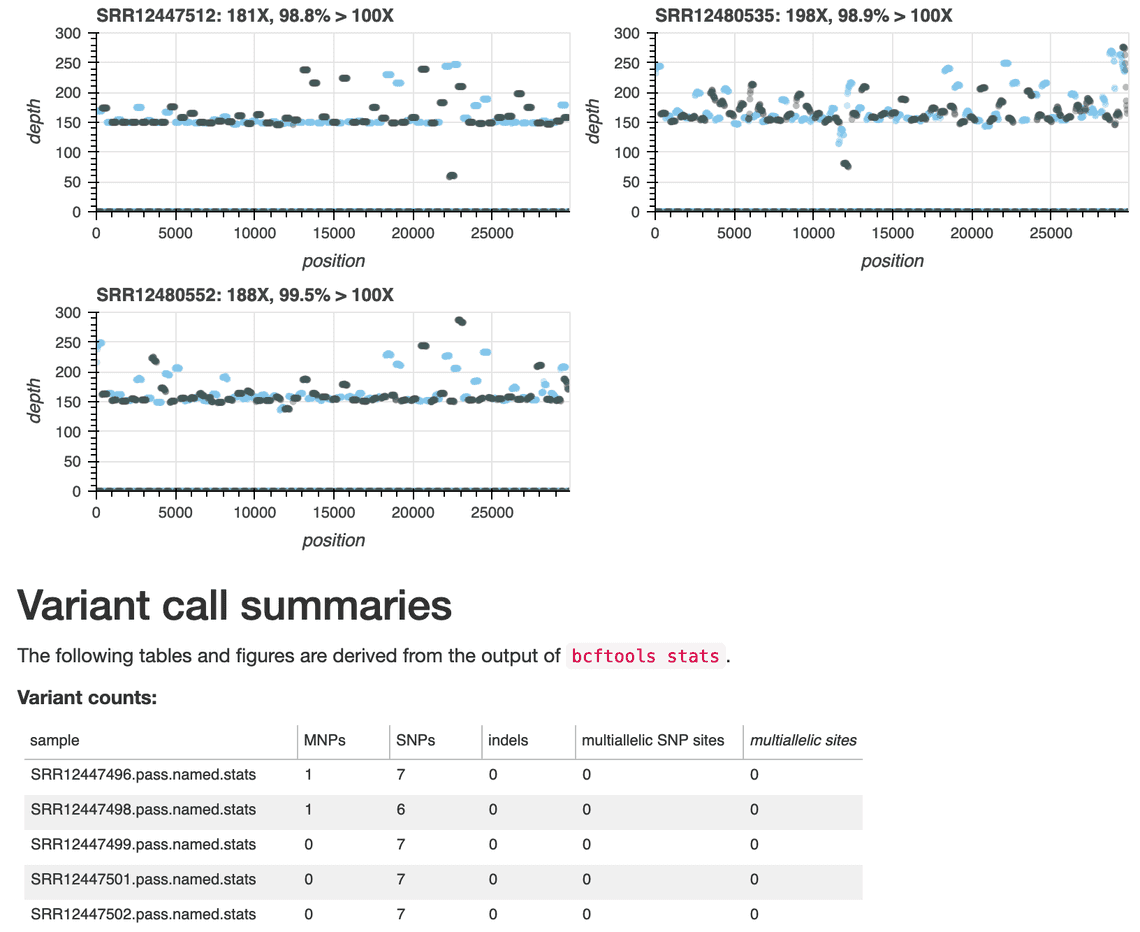
Figure 1. An excerpt from the wf-artic report; the information shown in the plots can be used to assess depth-of-coverage across the sequenced primer sets and to identify primers and amplicons that may have dropped out of the analysis. Additional data and their provenance are shown to further summarise the data and SARS-CoV-2 variants contained within. The dataset summarised in this sample report is from sequencing study PRJNA650037.

