Input directory structure for EPI2ME workflows

The EPI2ME team strives to make bioinformatics analysis as easy to use and intuitive as possible. Our nextflow workflows are designed to seamlessly run with Oxford Nanopore Technologies’ device outputs. They accept sequencing data in the format output by the sequencer, as well as forms into which users may have pre-processed their data. In this blog post we will explore the options provided and note a few minor exceptions to the rule.
So, without further ado, let’s dive into the dos and don’ts of passing data to one of our workflows.
Input file types
All EPI2ME workflows accept FASTQ files and an increasing number can also be used with BAM.
On the command line, you would pass FASTQ input with the --fastq flag and BAM with --bam.
In EPI2ME Desktop, the corresponding options are in the Input Options panel when launching a new workflow (see Figure below).
The expected file extensions are [.fastq, .fastq.gz, .fq, .fq.gz] and [.bam, .ubam], respectively.
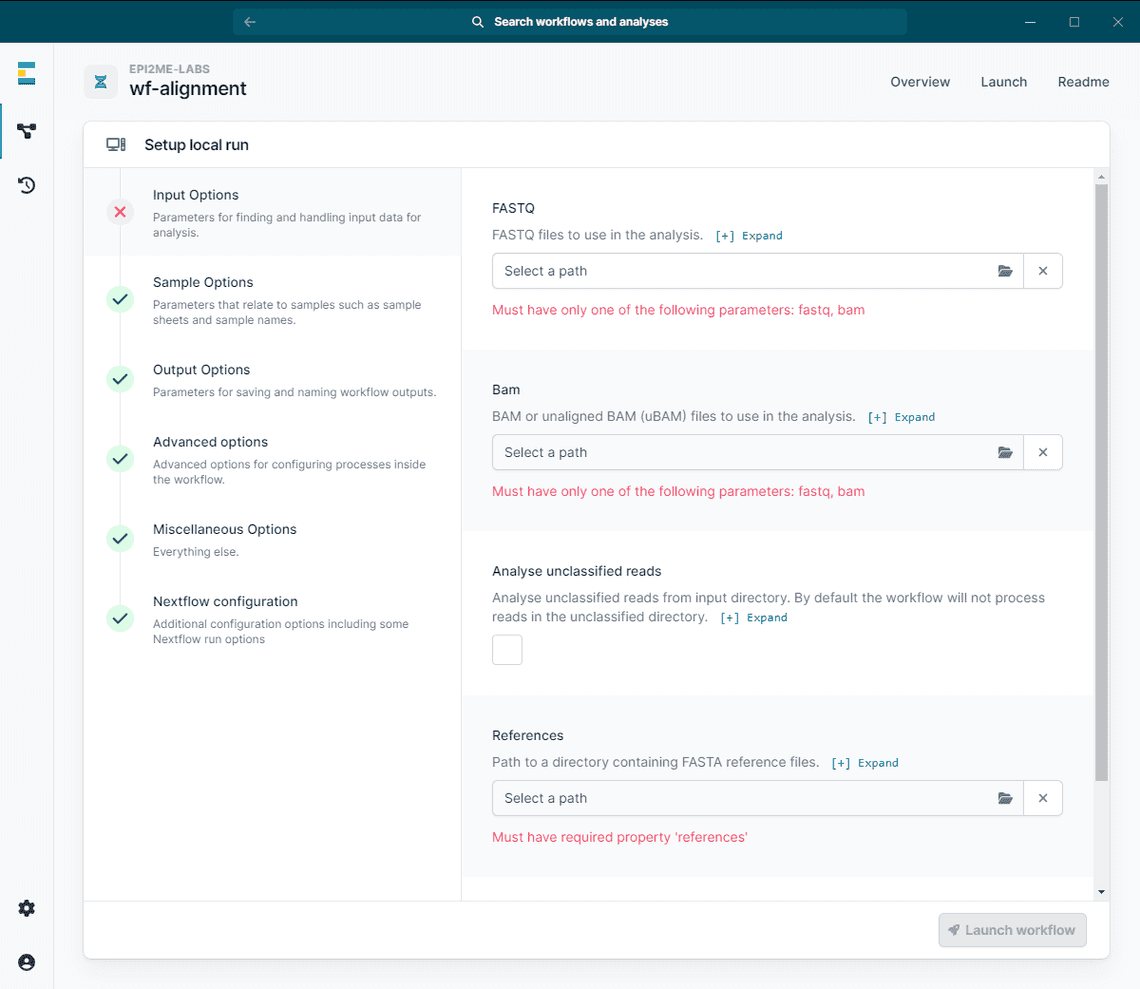
Input structure
Whether in FASTQ or BAM format, input data for our workflows should be structured in one of these three ways:
- a single file,
- a single directory containing several target (e.g. FASTQ) files, or
- a directory containing sub-directories (usually barcodes) which in turn contain the target files.
Some exceptions to these options are noted below.
Example file trees for the three cases might look as follows:
# single file # single directory # barcoded directories. . .└── reads.fq.gz └── my-input-dir └── my-input-dir├── reads1.fq.gz ├── barcode01├── reads2.fq.gz │ ├── reads1.fq.gz└── reads3.fq.gz │ ├── reads2.fq.gz│ └── reads3.fq.gz├── barcode02│ ├── reads1.fq.gz│ ├── reads2.fq.gz│ └── reads3.fq.gz├── barcode03│ ├── reads1.fq.gz│ ├── reads2.fq.gz│ └── reads3.fq.gz├── ...│...│└── unclassified├── reads1.fq.gz├── reads2.fq.gz└── reads3.fq.gz
In the third case, the sub-directories can only contain target files “one level deep”. In other words, the following is not allowed:
# this will fail input validation.├── barcode01│ ├── dir-within-barcode│ │ └── reads.fq.gz│ ├── reads1.fq.gz│ ├── reads2.fq.gz│ └── reads3.fq.gz...
Similarly, “mixtures” of the second and third case (i.e. target files in the top-level directory and in barcodes) will also fail:
# this will fail input validation.├── reads.fq.gz├── barcode01│ ├── reads1.fq.gz│ ├── reads2.fq.gz│ └── reads3.fq.gz...
The workflows are inflexible in this way such that they are able to correctly interpret the users intentions as to which files should be used for analysis.
Non-target files and directories containing only such files will be ignored. The below is therefore allowed:
# this will ignore non-accepted input files.├── some-other-file.txt├── barcode01│ ├── reads1.fq.gz│ ├── reads2.fq.gz│ ├── reads3.fq.gz│ └── yet-another-file.csv...
Exceptions
Most of our workflows behave precisely as above. Some, however, deviate and are more strict or accept only a subset of the options. The workflows wf-human-variation and wf-somatic-variation handle a single sample only; the options of directories of samples are therefore not relevant. By contrast, wf-tb-amr relies on the use of control samples and therefore explicitly requires multiple samples and a sample sheet (see below). Please refer to the respective READMEs for more details regarding their input data requirements.
Unclassified reads
The unclassified directory contains reads for which no barcode could be assigned during demultiplexing and it is ignored by default.
It can be included in the analysis with the --analyse_unclassified flag.
In EPI2ME Desktop, this parameter is usually found in the Input Options panel (just like the options for --fastq and --bam).
Sample sheets
When using barcoded directories, a sample sheet with metadata for the individual barcodes can be provided. This needs to be a CSV file with at least the following two columns (extra columns are allowed):
alias: The alias (or sample name) for a barcode / directory.barcode: The barcode. This needs to be in the formatbarcode(X)YZwith(X)YZdenoting either two or three integers (e.g.barcode01orbarcode001). All values in this column need to be of the same length (i.e. usingbarcode001as well asbarcode02in the same sample sheet is not allowed).
Some workflows require additional columns.
wf-tb-amr, for example, also expects a type column. (accepted values are test_sample, positive_control, negative_control, and no_template_control]).
A sample sheet with four samples might look like this:
barcode,alias,typebarcode01,sample1,test_samplebarcode02,sample2,test_samplebarcode03,positive,positive_controlbarcode04,negative,negative_control
Note: The above follows the MinKNOW sample sheet specifications and thus a valid MinKNOW sample sheet will also work with EPI2ME workflows.
Special characters
Whitespace and special characters in sample names, file paths, or reference sequence IDs can cause certain bioinformatics tools to fail.
EPI2ME workflows contain precautions to prevent this from happening, but we still recommend to only use alphanumerical characters (i.e. numbers and lower- or upper-case letters from ‘a’ to ‘z’) and underscores.
For example, sample_01 is fine, but sample_A*2:01/10 is not.
Real-time analysis with --watch_path
At the time of writing, wf-metagenomics and wf-basecalling can be run in real time.
To enable this, the flag --watch_path needs to be provided and the input cannot be a single FASTQ / BAM file.
Nextflow will then watch the input directory for new files with the correct extensions to appear and the pipeline will analyse them individually whenever they become available.
Summary
We hope the above was helpful and clarified how to provide input data to EPI2ME workflows. To recap:
- EPI2ME workflows accept either FASTQ and / or BAM input files which can be structured in one of three ways (a single target file, a single top-level directory containing target files, a directory containing barcoded sub-directories with target files).
- “Mixing” of these structures is not allowed (i.e. you cannot use a directory that contains barcodes as well as target files).
- Unclassified reads are ignored by default, but this behaviour can be overridden.
- Additional metadata (like sample names) can be provided with a sample sheet.
- Some workflows allow analysis of input files in real time (i.e. as they are being created by the sequencing device).
- Special characters should be avoided as much as possible.
When keeping these points in mind, analysing your data should be as easy as selecting the path to the inputs and pressing a button.
If you have further questions or run into issues, please let us know.
Further information
- An introduction to SAM and BAM files.
- FASTQ format
- SAM format
- BAM format
- EPI2ME Desktop quickstart
- Transition from EPI2ME Agent to EPI2ME Desktop
Share
Julian Libiseller-Egger
Bioinformatician
Table Of Contents
Related Posts
Information
