Phased CpG Methylation Calling in GM24385 with Remora and Clair3
Previously we made available an Oxford Nanopore Open Data release comprising 5-methylcytosine basecalls using the Guppy basecaller tailored to the specific task of identifying 5mC. In this post we present fresh set of basecalls using the new algorithms for the Remora project, and integrated into the research-grade basecaller Bonito.
For more information and help downloading data from our open dataset archive see the Datasets Tutorials page. All the data referred to in this blog can be accessed under:
s3://ont-open-data/gm24385_mod_2021.09/extra_analysis/bonito_remora
The most relevant files stored under this top level and referred to below are:
240.6 GiB all.bam76.9 MiB all.bam.bai233.6 GiB all.hp.bam76.9 MiB all.hp.bam.bai75.2 MiB all_contigs.vcf.gz1.5 MiB all_contigs.vcf.gz.tbi891.4 MiB bonito.cpg.bed.gz1.9 MiB bonito.cpg.bed.gz.tbi727.9 MiB bonito.hp1.cpg.bed.gz1.8 MiB bonito.hp1.cpg.bed.gz.tbi722.6 MiB bonito.hp2.cpg.bed.gz1.8 MiB bonito.hp2.cpg.bed.gz.tbi
Please also refer to the original post introducing this dataset.
The GM24385 cell line samples were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research.
Bonito-Remora Basecalling
Previous iterations of modified-base basecalling in the Guppy basecaller have required use of a basecalling model specific to the task of identifying modified bases. These models have typically traded the ability to call base modifications for a slight decrease in canonical basecalling accuracy. The advent of the new algorithms of Remora allow highest accuracy basecalling and identification of modified bases in a single basecalling process, reducing the computation requirements to obtain such results.
As with the analysis workflow using Guppy, the Bonito basecaller is capable of outputting
BAM files annotated with methylation calls as described in the
SAM tags specification found at: https://samtools.github.io/hts-specs.
Bonito can be provided with a reference genome and instructed to output BAM files with the MM
and ML tags described in the specification documents:
bonito basecaller dna_r9.4.1_e8_sup@v3.3 \<fast5 input directory> \--modified-bases 5mC \--reference <minimap2 reference index> \--recursive \--alignment-threads 8 \| samtools view -u | samtools sort -@ 8 > <output.bam>samtools index <output.bam>
Similarly the resultant BAM file can be summarized to a bedMethyl using the modbam2bed program (available through conda for both Linux and MacOS):
modbam2bed \-e -m 5mC --cpg -t 10 \GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \<bonito bams> ... \| bgzip -c > bonito.cpg.bed.gz
We therefore have obtained highest accuracy basecalls and modification calls in a useful summarised form with only two computational steps from the primary sequencer measurements.
Phased Methylation Calls
To further demonstrate the enhanced utility of long-read nanopore sequencing for modified-base
identification, we are also providing phased methylation calls. These have been produced de-novo
using our wf-human-snp Nextflow workflow to produce phased small variant calls
using clair3 and whatshap.
The phased variants were used to tag reads as belonging to one of the two haplotypes, and modbam2bed
run to produce a bedMethyl file per haplotype.
The variant calling workflow was run using,
nextflow run epi2me-labs/wf-human-snp \-r v0.1.2 --model r941_prom_sup_g5014 --phase_vcf \--bam all.bam \--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \--out_dir clair3 -w clair3
to produce a VCF file containing phased variants. These phased variants were then used
to tag each read as belonging to one of the two haplotypes, whatshap was used for this
task with,
whatshap haplotag \--ignore-read-groups \--output all.hp.bam \--reference GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \all_contigs.vcf.gz all.bam
The above command produced a BAM file with an HP (haplotype) tag for each read. Phased methylation
statistics were obtained using modbam2bed with the --haplotype option, once for each
haplotype:
for HP in 1 2; domodbam2bed \-e -m 5mC --cpg -t 10 --haplotype ${HP} \GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \all.hp.bam \| bgzip -c > bonito.hp${HP}.cpg.bed.gzdone;
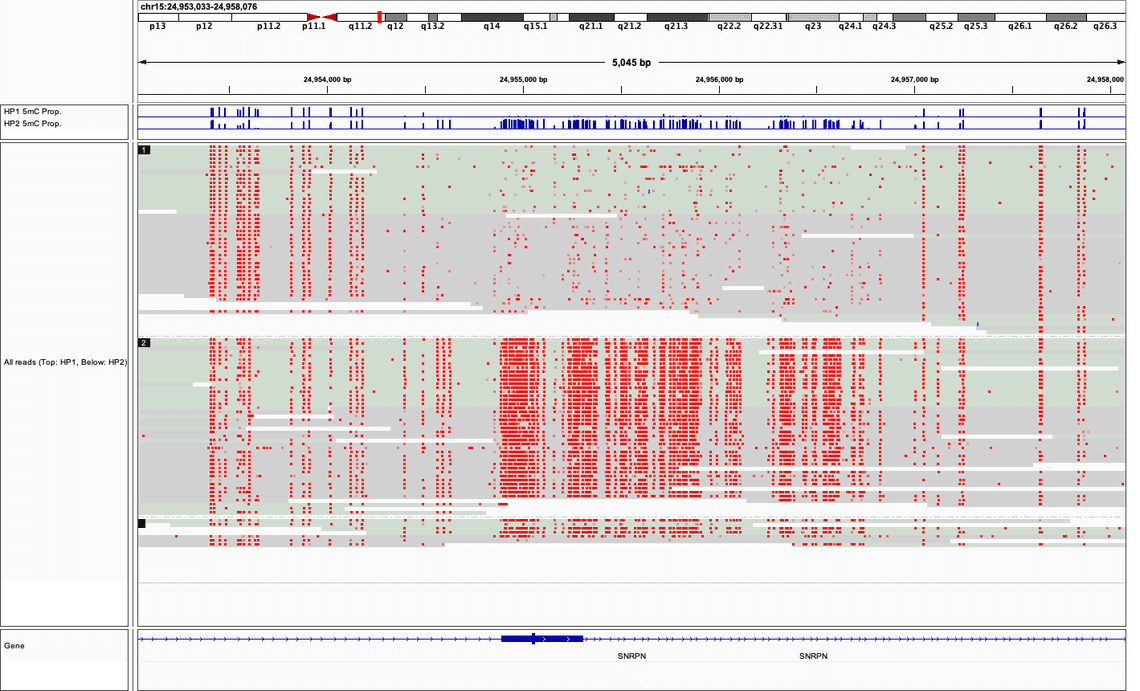
As a well-known and characterised example of differential methylation between maternal and paternal haplotypes, Figure 2. depicts sequencing data in the region around the Prader-Willi Syndrome associated gene SNRPN. Using the procedure above sequencing data is tagged as either “haplotype 1” or “haplotype 2”: a striking difference in the rate of 5mC presence is observed.
Discussion
Here we have shown how the latest software tools from Oxford Nanopore Technologies can be used to obtain simply phased CpG modification calls for the GM24385 human cell-line. The methods used are applicable to any diploid sample. We hope that these new tools will greatly accelerate fields of research where DNA methylation is known to play an important role, and also unlock new insights.

