Structural variation calling with GM24385
In this blog post we will explore structural variant calling using the recently released HG002 (GM24385 Ashkenazi Son) data release.
The GM24385 dataset comprises whole genome sequencing of a well-characterised human cell line. It therefore provides a useful benchmark sample; the cell line was also used as a “seen” sample in the recent PrecisionFDA Truth Challenge V2 competition for small variant calling.
Structural variant calling with lra and cuteSV
As an easily reproducible example we will focus on a single flowcell of the GM24385 2020.11 data release.
This walkthrough assumes some familiarity with standard bioinformatic tools for handling genomics data. A working installation of samtools, snakemake, git, and the AWS command-line tools are required to follow the process below.
Data preparation
We will start by downloading Guppy 4.0.11 basecalls from a PromethION sequencing experiment (see our tutorial FAQs) for more information on downloading data):
aws s3 --no-sign-request cp \s3://ont-open-data/gm24385_2020.11/analysis/r9.4.1/20201026_1644_2-E5-H5_PAG07162_d7f262d5/guppy_v4.0.11_r9.4.1_hac_prom/basecalls.fastq.gz \basecalls.fastq.gz
The .fastq file downloaded above contains the QC pass calls from the experiment
amounting to around 200 Gbases.
To run the SV calling pipeline and perform benchmarking we will need release 37 of the human reference sequence:
wget http://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/human_g1k_v37.fasta.gz
Running the variant calling
To perform structural variant calling Oxford Nanopore Technologies recommends using the pipeline-structural-variation snakemake workflow. This workflow as been recently updated to use lra and cuteSV, replacing the previous minimap2 and sniffles based approach. After installation of this software we use it with its default settings:
conda activate pipeline-structural-variation-v2snakemake call --config \input_fastq=basecalls.fastq.gz \reference_fasta=human_g1k_v37.fasta \threads=76 \sample_name=PAG07162-hg37
The useful output for our purposes is the single Variant Call Format file; a copy of the file is available in the dataset S3 bucket at:
s3://ont-open-data/gm24385_2020.11/extra_analyses/sv_calling/PAG07162-hg37/sv_calls/PAG07162-hg37_cutesv_filtered.vcf.gz
Evaluation
The veracity of the variant calling performed above can be obtained by comparing the results to the Genome In A Bottle truth sets for the GM24385 sample. The truth sets can be downloaded from the NCBI repository:
truth_name="HG002_SVs_Tier1_v0.6"for ext in ".vcf.gz", ".vcf.gz.tbi", ".bed"; dowget -O $truth_name$ext \https://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/NIST_SVs_Integration_v0.6/$truth_name$extdone
With these reference data we will use truvari to assess the recall and precision of the variant calls made by the calling pipeline:
truth_vcf="${truth_name}.vcf.gz"truth_bed="${truth_name}.bed"reference="human_g1k_v37.fasta"input_vcf="PAG07162-hg37_cutesv_filtered.vcf.gz"output_dir="truvari"truvari bench --passonly --pctsim 0 \-b $truth_vcf --includebed $truth_bed \-f $reference -c $input_vcf \-o $output_dir
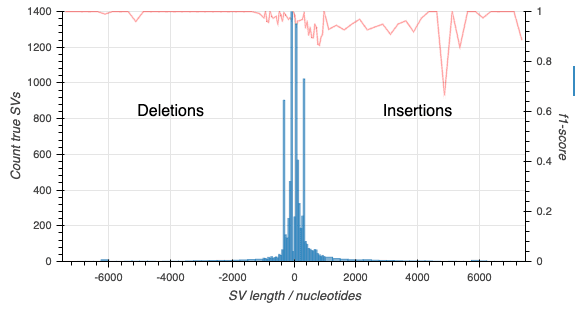
Truvari outputs precision and recall figures for the structural variants. With a little work (detailed in the EPI2MELabs Structural Variation Benchmarking tutorial) we can separate the counts for deletion and insertion (including duplication) variants:
| Type | Deletions | Insertions |
|---|---|---|
| Recall | 0.9667 | 0.9427 |
| Precision | 0.9893 | 0.9667 |
| f1-score | 0.9779 | 0.9545 |
With still a little more work we can produce the following depicting the f1-score alongwith counts of SVs in the truthset:
Related Posts
Information

