Nextflow and Jupyter Labs
We are pleased to introduce an update to our EPI2ME Labs product that now includes the functionality to launch our Nextflow pipelines directly from the EPI2ME Labs session. These new capabilities have been implemented to facilitate the launching of integrated workflows in an efficient manner whilst avoiding the requirement for using a terminal session.
Getting started
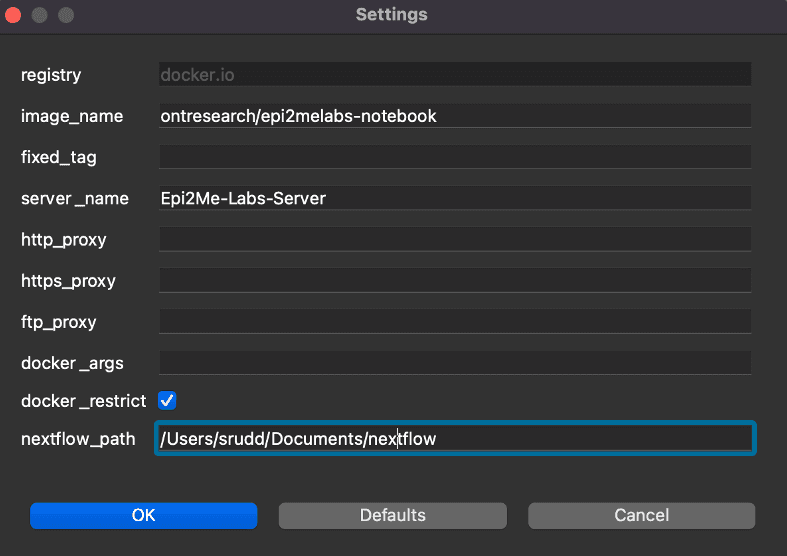
Install the new version of the EPI2ME Labs Launcher; this can be downloaded from our Downloads page. The LabsLauncher interface is largely unchanged – there is one small, but critical, requirement – we need to specify the path to the Nextflow binary.
The new EPI2ME Labs landing page
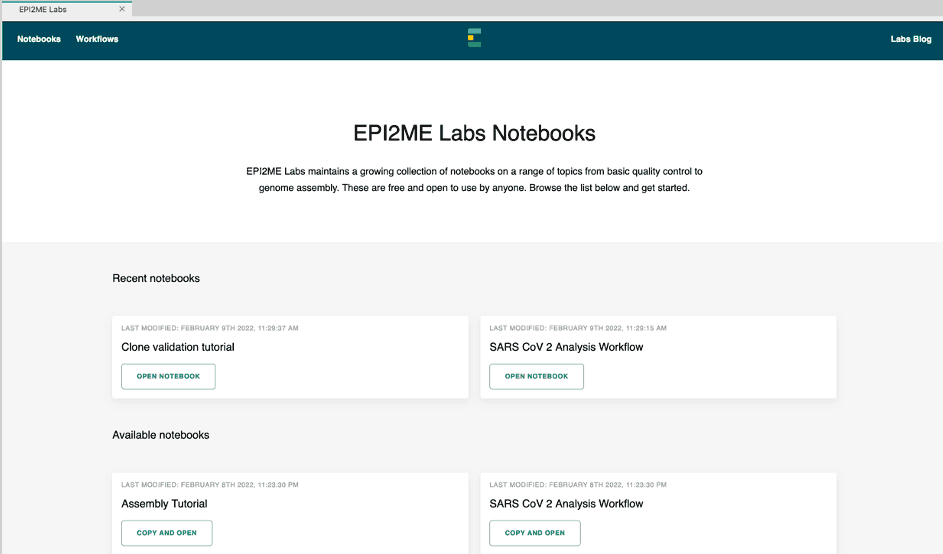
When EPI2ME Labs is launched a new landing page is now presented. The landing page still provides the list of recently opened and available tutorials but now also provides a link to the EPI2ME Labs Workflows. The landing page is shown in Figure 2.
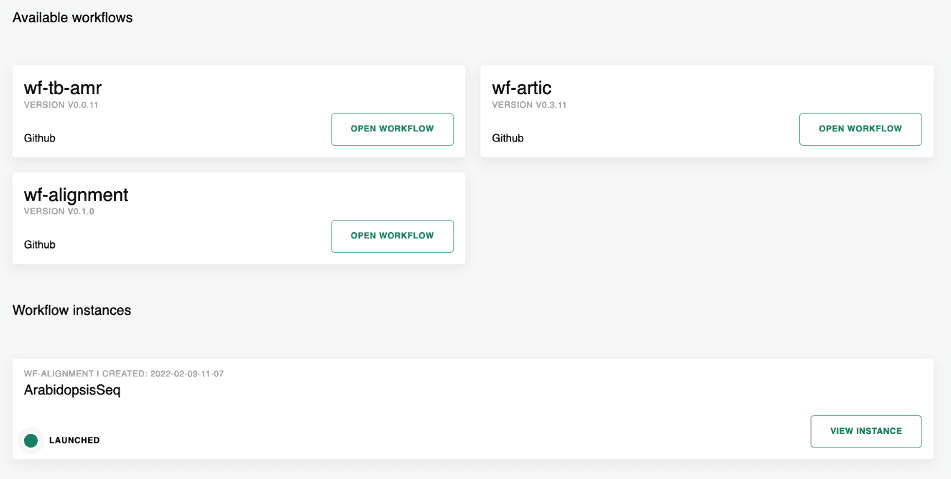
When the “Workflows” link is selected from the navigation bar at the top of the landing page, the available workflows will be presented. The workflows page also shows the list of workflows that have been run, or are still running. Figure 3 shows a screenshot of the Workflows page – this shows three of the available workflows that may be selected and shows that one workflow, ArabidopsisSeq is currently running.
Running a workflow
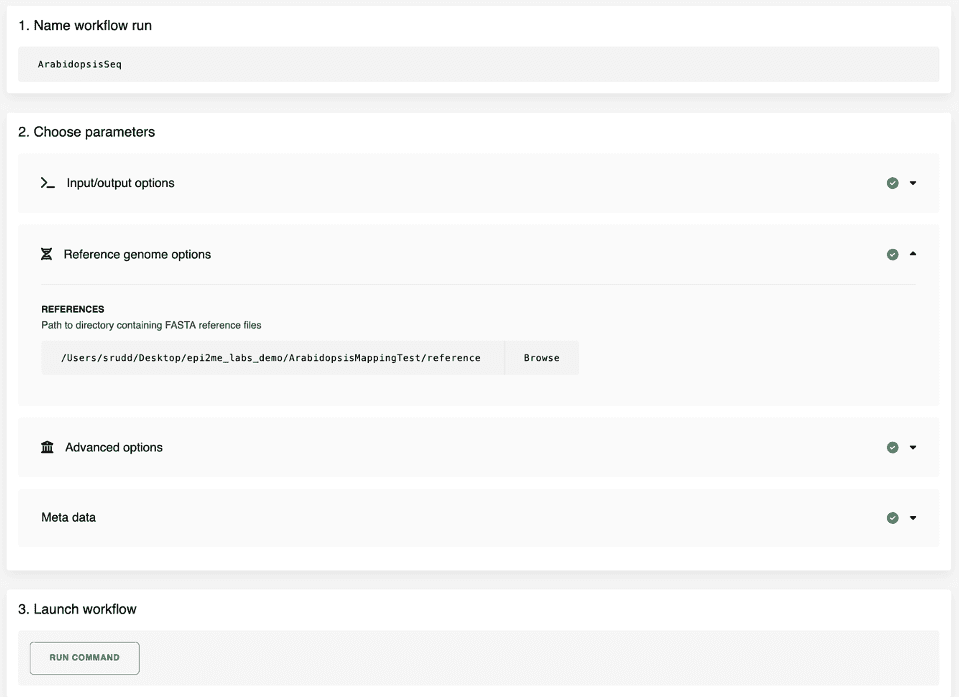
To run a workflow is now simple: select a workflow from the list of available workflows. This will open a new dialog page that will encourage you to specify a nextflow analysis. Figure 4 shows how an analysis can be built for the wf-alignment workflow – we need to specify both the collection of FASTQ files to map and the FASTA format reference genome. The analysis is started with a click of the “RUN COMMAND” button and the analysis will commence.
Once a workflow has been started the experiment information becomes available as a workflow instance. Each instance has its own page and this provides both the output from the Nextflow command, links to the result files and additional metadata on the version of the workflow called, the parameters passed and the actual command run on the computer. Figure 5 shows an example of a workflow instance screen for the wf-alignment workflow that we have building.
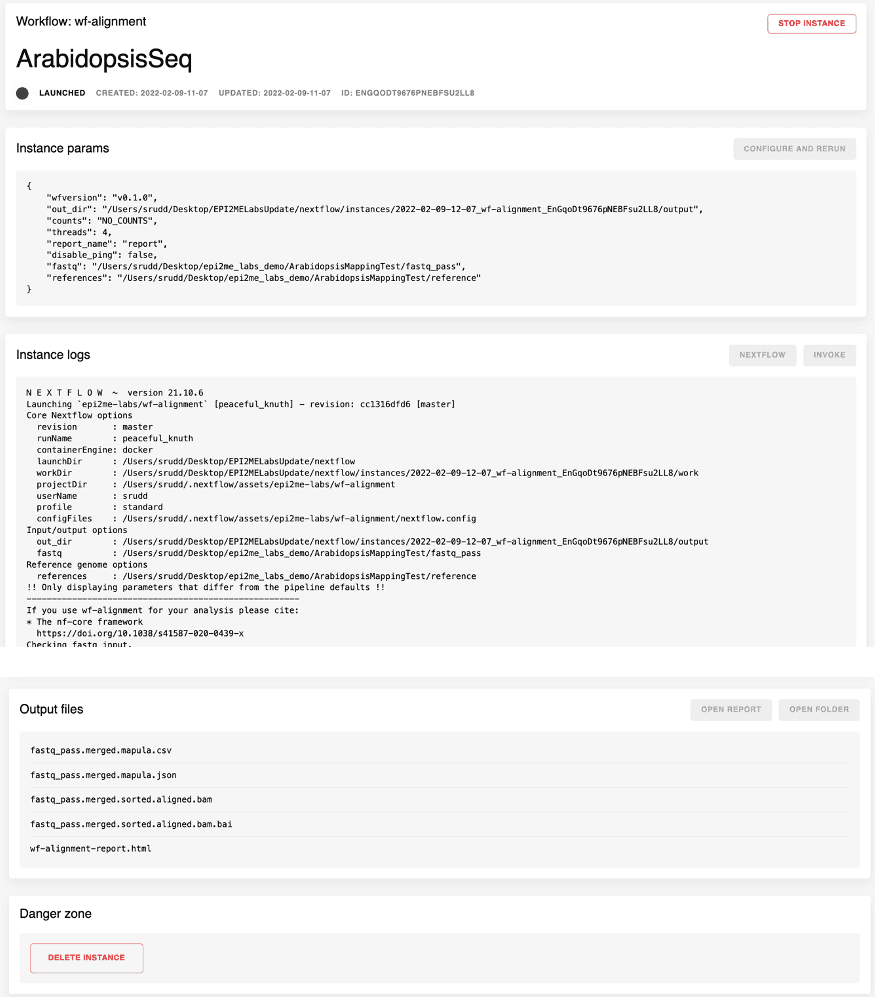
When the analysis has completed the results are made available through the same instance report page. The analysis report is accessible by clicking on the “OPEN REPORT” button.
Share
Related Posts
Information

