Modified Base Best Practices and Benchmarking

Modified bases, including methylation, regulate many biological processes - from eukaryotic gene expression to bacterial immunity. Methylation plays a pivotal role in human health, influencing cancer development, neurological disorders, cardiovascular diseases, and other conditions through the regulation of cellular processes.
In the context of nanopore sequencing, modified bases can be detected and distinguished through perturbations to the measured ionic current. These differences are exploited in basecalling, but can also be leveraged in more detailed analyses.
In this post we will outline best practices for performing modified base detection with nanopore sequencing, and present high accuracy benchmark results for three common DNA methylation marks:
- 5-methylcytosine (5mC)
- 5-hydroxymethylcytosine (5hmC)
- 6-methyladenine (6mA)
Benchmarking results are derived from synthetic oligonucleotides, each containing canonical (unmodified) or modified bases within all distinct 5-mer sequence contexts. We also provide raw data, tools, and step-by-step instructions for running a validation pipeline to replicate these results.
Data Access
Raw nanopore data for canonical and modified samples, reference sequences, and annotations of canonical and modified positions are available for download.
These datasets allow users to follow the analyses described in this post or expand upon them to conduct more in-depth investigations. We include two datasets: a “full” dataset and a “subset” dataset. Both are provided as raw sequencer outputs in pod5 format, ideal for signal visualization with Remora or custom signal processing algorithm development. The full dataset comprises all data that was collected during experimentation. The subset dataset was produced from the full dataset by aligning to the provided reference sequences randomly selecting 5,000 reads per synthetic construct. The reference-balanced subset is intended to allow users to quickly reproduce results and investigate the synthetic datasets. Provided basecalls allow users to inspect modified base calls without the needing to run the basecalling step described below. Basecalling for those BAMs provided were performed on the subset dataset with the SUP basecallling model.
The data is located on AWS S3 at:
s3://ont-open-data/modbase-validation_2024.10/
The structure of the S3 prefix is shown below.
.├── full| ├── control_rep1.pod5| ├── control_rep2.pod5| ├── 5mC_rep1.pod5| ├── 5mC_rep2.pod5| ├── 5hmC_rep1.pod5| ├── 5hmC_rep2.pod5| ├── 6mA_rep1.pod5| └── 6mA_rep2.pod5├── subset| ├── control_rep1.pod5| ├── control_rep2.pod5| ├── 5mC_rep1.pod5| ├── 5mC_rep2.pod5| ├── 5hmC_rep1.pod5| ├── 5hmC_rep2.pod5| ├── 6mA_rep1.pod5| └── 6mA_rep2.pod5├── basecalls| ├── control_rep1.bam| ├── control_rep2.bam| ├── 5mC_rep1.bam| ├── 5mC_rep2.bam| ├── 5hmC_rep1.bam| ├── 5hmC_rep2.bam| ├── 6mA_rep1.bam| └── 6mA_rep2.bam└── references├── all_5mers.fa├── all_5mers_C_sites.bed├── all_5mers_A_sites.bed├── all_5mers_5mC_sites.bed├── all_5mers_5hmC_sites.bed└── all_5mers_6mA_sites.bed
For more information and help downloading data from our open dataset archive, see the Datasets Tutorials page. Analyses based on these data are presented below.
Why Synthetic Datasets?
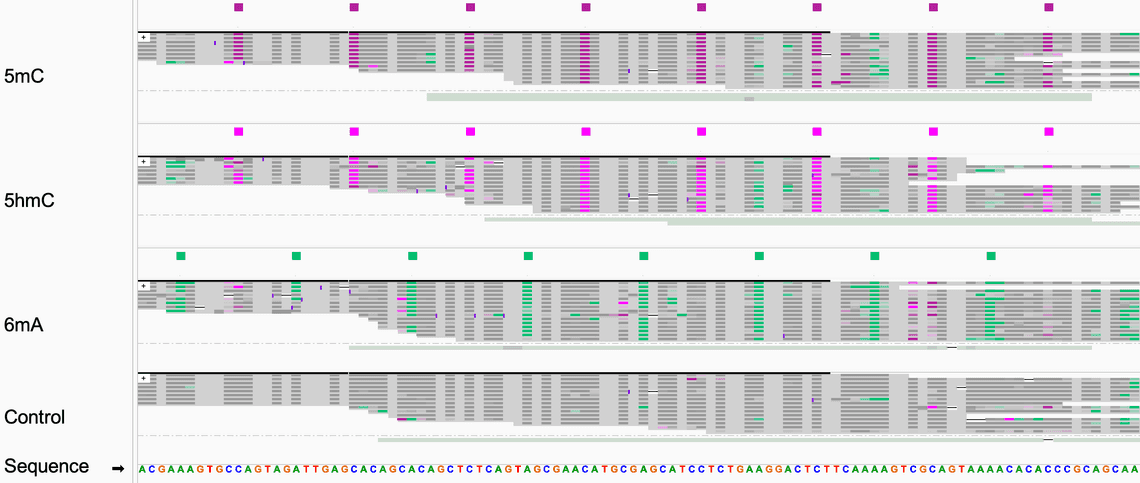
Validating modified base models at single-molecule and single-base resolution is challenging due to the complexity of identifying reliable ground truth datasets. At Oxford Nanopore Technologies, we produce synthetic oligonucleotides to obtain the highest quality validation data for model evaluation. The validation dataset for each modified base includes oligonucleotides covering all possible 5-mer sequence contexts. A depiction of the sequencing reads is shown in Fig. 1. The validation dataset was sequenced on a PromethION-24 device.
Best Practices for Basecalling and Modified Base Validation Analysis
This tutorial uses two open source tools available on GitHub: Dorado for basecalling, including modified base calling, and Modkit for validating modified base calls. Both are command-line tools from Oxford Nanopore Technologies. While we use specific versions in this tutorial, we strongly recommend using the latest releases of both tools for optimal performance and accuracy.
Setting Up Your Tools
To perform the analysis we need to first install our two tools, both are available as pre-compiled binaries in tar archives.
Install Dorado (v0.8.2 used here):
wget https://cdn.oxfordnanoportal.com/software/analysis/dorado-0.8.2-linux-x64.tar.gz -O - | tar -xzInstall Modkit (v0.4.1 used here):
wget https://github.com/nanoporetech/modkit/releases/download/v0.4.1/modkit_v0.4.1_u16_x86_64.tar.gz -O - | tar -xz
Running a Validation Analysis
The first step of our analysis is to perform basecalling from the raw sequencer outputs.
This is done with the dorado basecaller, for example using the control_rep1 dataset:
# step 1: running basecalling with doradodorado basecaller \hac,5mC_5hmC,6mA \subset/control_rep1.pod5 \--reference references/all_5mers.fa \> control_rep1_basecalls.bam
The command will generate a BAM file containing both standard basecalls and modified base information stored in community standard BAM tags.
The command can be repeated for the 5mC_rep1 and 5hmC_rep1 datasets
We can now use Modkit to validate modified base calls from the control and modified datasets:
# step 2: validate results with modkitmodkit validate \--bam-and-bed control_rep1_basecalls.bam references/all_5mers_C_sites.bed \--bam-and-bed 5mC_rep1_basecalls.bam references/all_5mers_5mC_sites.bed \--bam-and-bed 5hmC_rep1_basecalls.bam references/all_5mers_5hmC_sites.bed \--min-identity 10 \--out-filepath validate_5mC_5hmC_mods.txt \--log-filepath validate_5mC_5hmC_mods.log
This command requires data with known modified base content at each position of each read, such as the synthetic oligonucleotide datasets provided here.
The modkit validate command automatically filters the modified base calls using a dynamic confidence threshold, retaining 90% of the data while optimising accuracy.
This approach balances precision (accuracy of the calls made) with recall (total number of calls), following standard machine learning practices.
The output provides detailed accuracy metrics for each model tested.
Benchmarking Modified Base Detection with Modkit
The modkit validate command is a versatile command when applied to different data types.
We can run the command on all applicable replicates with the appropriate ground truth files produce a variety of accuracy metrics. The table below depicts validation results for the following scenarios:
- Removing particular modified base types (for multiple modified base models)
- Limiting the sequence context within the ground truth strands
- Applied to different basecallers
| Modified Base | Context | HAC Accuracy | SUP Accuracy |
|---|---|---|---|
| 5mC+5hmC | All | 97.30 | 97.80 |
| 5mC+5hmC | CpG | 98.21 | 98.15 |
| 5mC only | All | 99.20 | 99.48 |
| 5mC only | CpG | 99.76 | 99.81 |
| 6mA | All | 96.21 | 97.60 |
Digging in a bit deeper, the HAC 5mC+5hmC calls validated on all-context sites results in the following confusion matrix (produced directly by modkit validate):
Falsely identified modified base calls are generally problematic for downstream pipelines.
Note that false modified base frequency (first row) is much lower than false canonical calls at modified sites.
Model training has been tuned to reduce false calls modified base calls for all models.
For applications requiring higher sensitivity, modkit provides options to adjusted filtering thresholds.
Isolating 5mC calls eliminates the possibility of misclassification between modified base types, resulting in higher accuracy (Fig. 3a). Many specific applications can be improved by limiting modified base calls to 5mC only. This process can be completed with the following Modkit command:
modkit \adjust-mods \--ignore h \control_rep1_basecalls.bam \control_rep1_basecalls_5mC_only.bam
Upgrading the basecalling model from high-accuracy to super-accuracy imparts a corresponding improvement in modified base accuracy (Fig. 3b). Analysis restricting to CG sequence contexts only, which are biologically important in many eukaryotic applications, shows a further refinement in accuracy (Fig. 3c). For the distinct modified base 6mA, we observe similar model characteristics with very low false modified base frequency (Fig. 4).
Discussion
These strands can also be used to more precisely specify expected results for modified base experiments. For example, these strands can be utilised to estimate the required coverage for specific tasks, such as differential methylation.
Additionally, these strands can help to calibrate expectations for applications of the many functions available from Modkit. Specifically, Modkit can perform the following common processing tasks:
- Differentially Methylated Region (
dmr) analysis (docs) localizemodified base content around genomic features of interest such as promoters or chromatin state (docs)entropyof methylation pattern (docs)- Exploring methylation motifs with the
motifcommands (docs)
For more examples of downstream modified base analyses with Modkit, see the Modkit poster.
In summary, this post provides a comprehensive guide to nanopore modified base analysis using synthetic ground truth data. By following the outlined best practices and leveraging the provided datasets, you can achieve accurate and reproducible modified calls, helping advance research in epigenetics and beyond.
