EPI2ME Labs 21.02 Release
We are delighted to release this month’s update to EPI2ME Labs. The 2021-02.01 release addresses some Community feedback and introduces new and updated tutorials.
ERCC spike-in quantification
A new tutorial to assess performance of a transcriptomics study through the use of spike-in controls.
Our ERCC tutorial introduces the synthetic sequencing controls developed by the External RNA Controls Consortium (ERCC). These spike-in control sequences are explored within an expression profiling study and demonstrate how the performance of a transcriptomics study may be benchmarked.
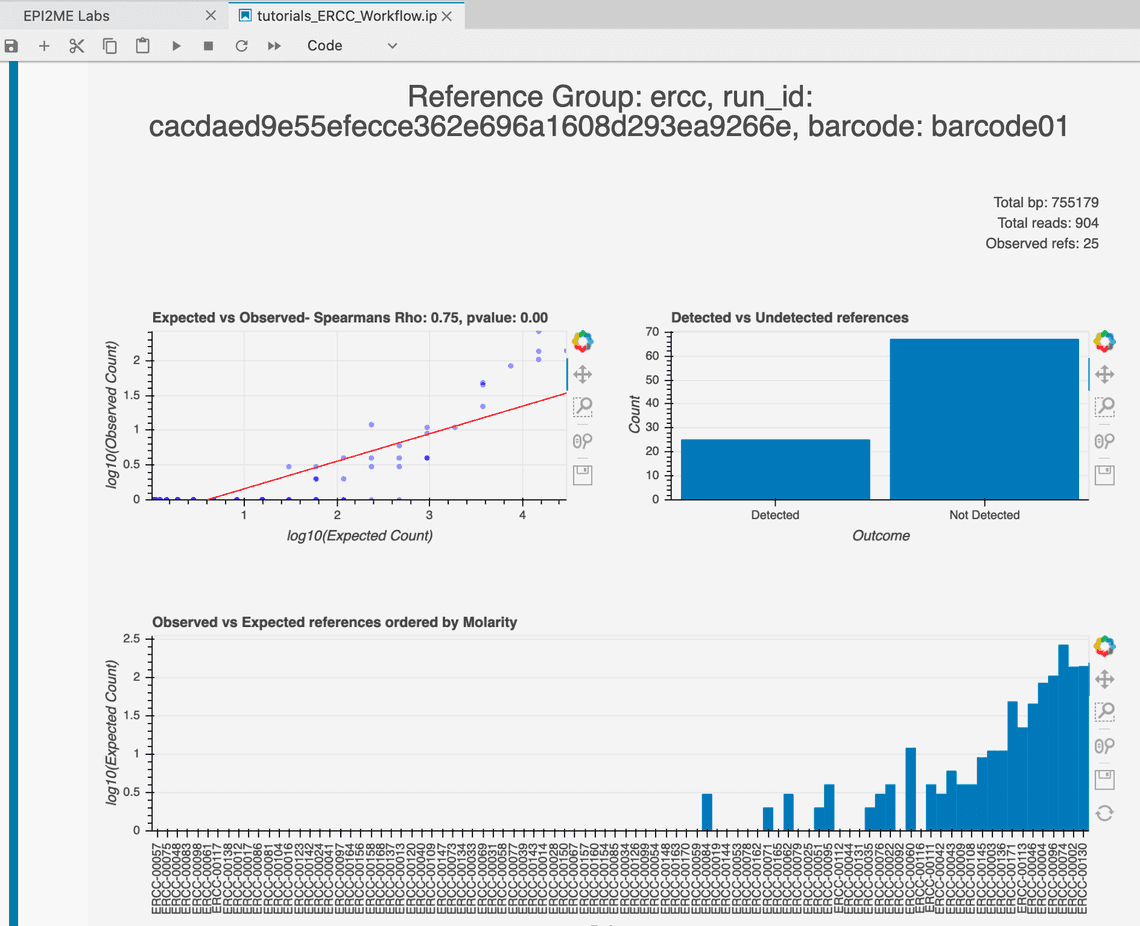
Figure 1. The ERCC tutorial introduces a workflow that looks for the presence of synthetic RNA transcripts and considers their observed abundance. The correlation between the observations of each ERCC with its expected counts can be used to assess study performance. The figure above shows some basic count measurements that are presented during the analysis.
SARS-CoV-2 ARTIC update
The SARS-CoV-2 tutorial introduces the ARTIC FieldBioinformatics workflow for the assembly and polishing of SARS-CoV-2 genomes. The workflow has been updated to demultiplex sequence collections using an expanded range of both native and rapid barcoding kits - the workflow has been tested with the “Eco” version of the PCR tiling of SARS-CoV-2 virus protocol. We appreciate that some users may prefer to perform demultiplexing during sequencing in MinKNOW; the tutorial now optionally processes FASTQ sequences that have already been demuliplexed. The tutorial includes a new and more interesting set of example data that have been picked from project PRJNA650037 at the European Nucleotide Archive.
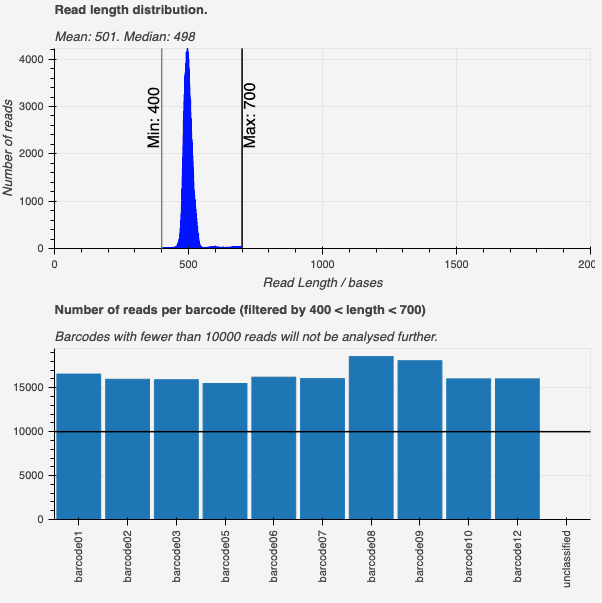
Figure 2. The SARS-CoV-2 ARTIC notebook walks users through the assessment of their ARTIC amplicon data to assess multiplexing efficiency. The final output of the workflow is a single FASTA formatted file containing a consensus sequence for each sample suitable for upload to GISAID or inspection with NextClade.
EPI2ME Labs Launcher support on more computer platforms
In addition to our previously packaged installations, we are now also packaging the EPI2ME Labs Launcher for Centos 7, Centos 8 and Ubuntu 20.04. The complete set of EPI2ME Labs Launcher software can be found at https://labs.epi2me.io/downloads/.
Command-line interface (CLI) to Labs Launcher
EPI2ME Labs has been installed on a variety of computer platforms. User feedback has revealed that there is interest in the installation of the EPI2ME Labs container on headless servers that offer greater memory and CPU resources. To support the installation of EPI2ME Labs on these more powerful computer systems, the EPI2ME Labs Launcher has been updated to include CLI functionality to start, stop, and update the appropriate services. More information on the CLI interface to the EPI2ME Labs Launcher can be found in a separate blog post.
Maintenance, updates and utility
The software environment in the epi2melabs-notebook has been updated to use Python version 3.8 (from Python 3.6). Housekeeping during the preparation of the docker container has considerably reduced the size of the container download - the download is now only approximately 1.3G in size. To increase the legibility of the functional code within the EPI2ME Labs tutorials and notebooks, we are introducing new software packages to simplify and structure analyses. aplanat has been used in earlier tutorials to simplify the preparation of figures - in this release we are introducing mapula to simplify the mathematics of processing read statistics and count information from BAM alignment files.
We look forwards to your feedback. We would welcome suggestions for future tutorials and we would gratefully review Community contributions.

