EPI2ME Labs 21.07.02 Release
Dear Nanopore Community,
Our (second!) July 2021 EPI2ME Labs release includes various updates to workflows and tutorials.
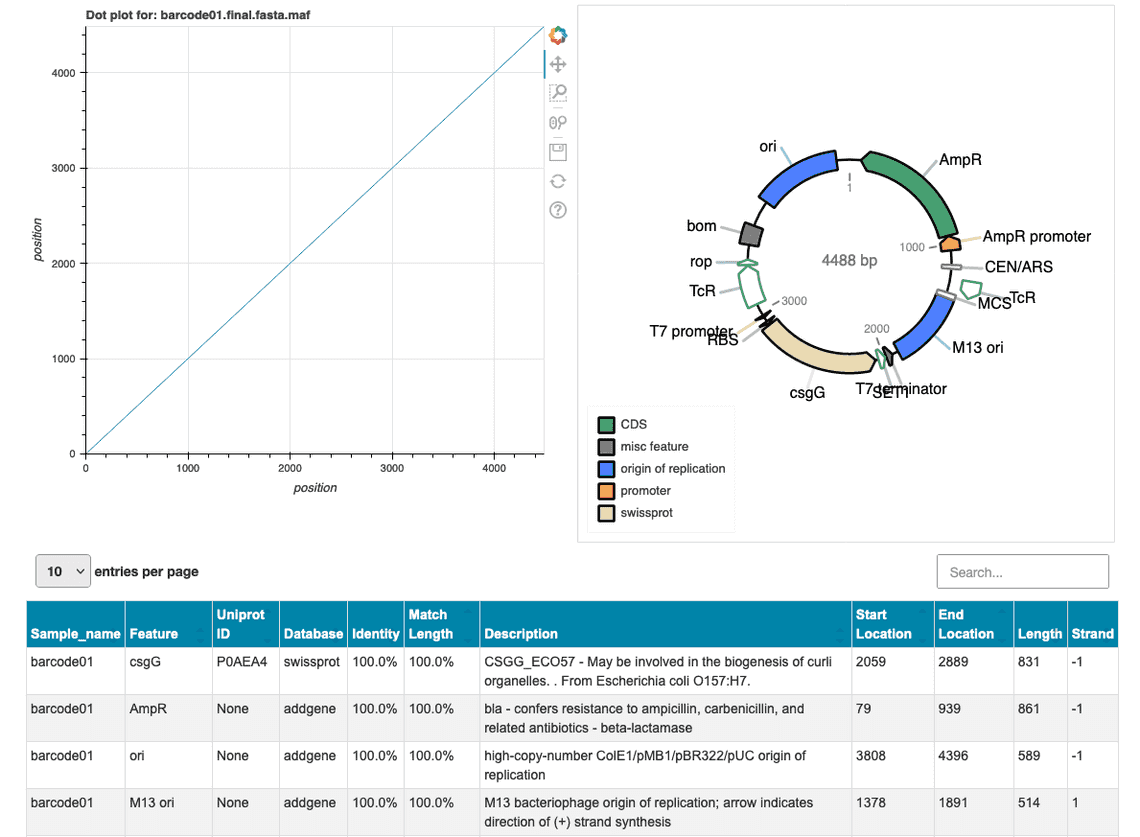
The wf-clone-validation workflow has been updated to version v0.1.0 and now includes the pLannotate tool for the visualisation and annotation of plasmid consensus sequences. The pLannotate software uses a collection of public sequence databases to explore the plasmid sequence for regions of interest that include e.g. the origin or replication and antibiotic resistance genes and may include information on the cloned insert sequence (if orthologues are present in the sequence database).
The wf-artic pipeline v0.3.1 release now reports a per-sample BAM file in the result directory. These BAM files are a standard component of the FieldBioinformatics workflow and correspond to the sorted sequence mapping data that has been structured by readgroup and trimmed of sequencing primers. This user request addresses the requirement that BAM files are also provided along with strain identification information.
The EPI2ME Labs SV tutorial has been updated to use LRA for read mapping and CuteSV for the identification of genomic structural variation; this synchronises the functionality between our research tool, pipeline-human-sv, and the EPI2ME Labs offerings. As a brief reminder, EPI2ME Labs provides a collection of best-practice tutorials that explore a variety of themes in the analysis of Nanopore sequence data. The EPI2ME Labs quick start guide provides more information.
The SARS-CoV-2 tutorial in EPI2ME Labs has been repaired - the V1200 primer set has been added back to the workflow.
We have made a number of minor changes to the way that we prepare and distribute our nextflow based workflows.
- The documentation for our 11 workflows (all are available through GitHub but a couple are still in development) now focuses only on the configuration, execution and results from the pipeline - we have removed the common introductory and configuration texts; these documents are provided in the quick start guide and additional information is also included in our how-to documents.
- We have improved our dockerfile housekeeping - this means that there are minor version updates to all our of workflows and the corresponding docker images are now considerably smaller.
- The environment.yaml files that define Conda channels and corresponding software requirements are now better defined in the nextflow.config. This update provides a more robust workflow experience for users who cannot use the recommended Docker software and/or prefer to run workflows inside a Conda environment (with the -profile conda option).
- Mamba is now included as our preferred software for the installation of software packages from Conda channels.
We look forwards to feedback and recommendations for future tutorials and workflows.

