SARS-CoV-2 Midnight Chemistry V14 Update

Introduction
Please note the
wf-articworkflow has now been updated to select the Medaka variant model based on the provided basecaller configuration (using the parameter--basecaller_cfg). Alternatively the Medaka model can be provided directly via the--medaka_variant_modelparameter.
We are pleased to offer a new version of wf-artic which supports models for optimal variant calling by medaka when using RBK114.96 and R10.4.1 flow cells for data generation.
This update coincides with the release of an early access protocol for Kit V14/R10.4.1 midnight on the Oxford Nanopore community which can be found here.
Versions of wf-artic >= v0.3.23 include this support for kit14/R10.4.1 medaka models. In this blog post we describe the improvements in Kit V14/R10.4.1 and how to run updated wf-artic with this data on the command line and EPI2ME Labs.
Why use Kit V14/R10.4.1 for SARS-CoV-2 sequencing?
Using the latest sequencing kits and flow cells from Oxford Nanopore Technologies ensures that you are getting the best possible data for your application. We recently released Kit V14 chemistry along with R10.4.1 flow cells which together deliver our most accurate sequencing data to date. This increase in accuracy is now available for our SARS-CoV-2 sequencing workflow, Midnight, using EXP-MRT001 and SQK-RBK114.96. We recommend users upgrade to get the highest accuracy Oxford Nanopore SARS-CoV-2 genomes.
For more information on the upgrade in performance see this post in the Oxford Nanopore community.
Alongside general accuracy improvements there are particularly impressive improvements in homopolymer regions when moving to Kit V14 and R10.4.1 flow cells. Figure 1 shows the proportion of non-deletion calls in an adenosine homopolymer in the spike gene of SARS-CoV-2 (taller blue bars mean better performance).
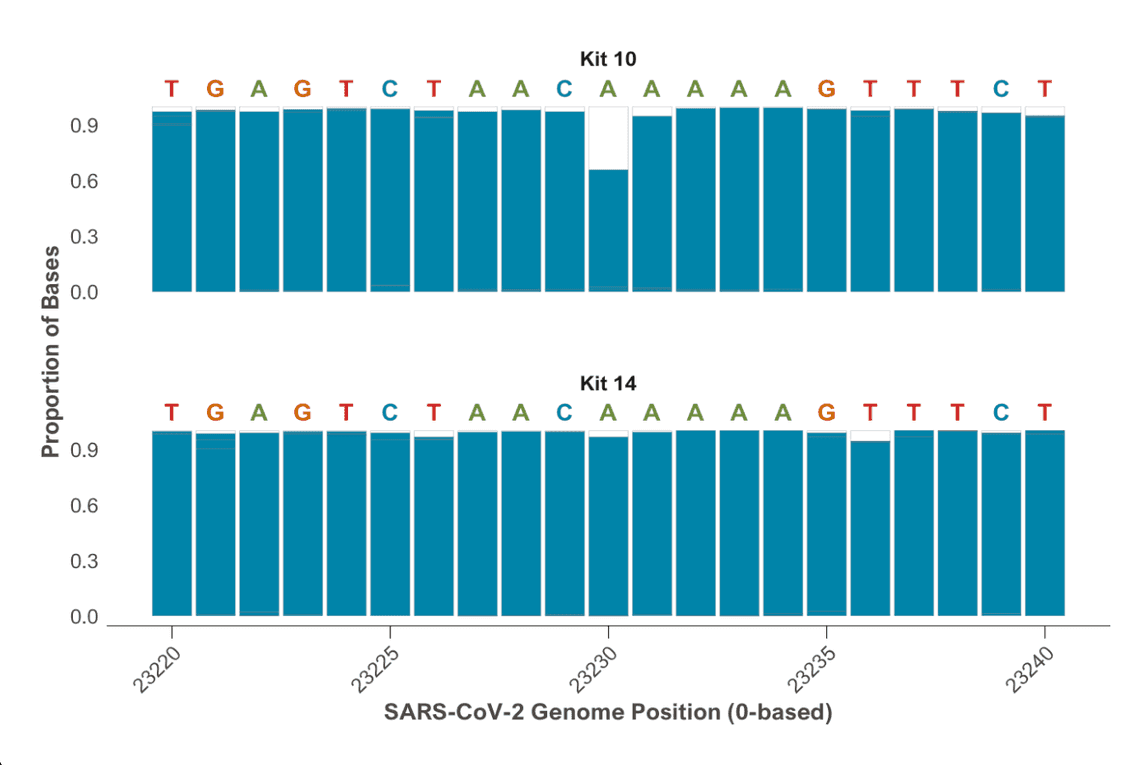
Analysis of Midnight Kit V14/R10.4.1 data
To take advantage of these improvements to our chemistry, medaka, which is used by the ARTIC Network workflow to call the variants in the SARS-CoV-2 genomic data, needed to be updated. This has now been completed and versions of wf-artic >= v0.3.23 include updated medaka which is able to use variant calling models best suited to Kit V14/R10.4.1 chemistry.
The new models included are:
r1041_e82_400bps_hac_variant_g615r1041_e82_400bps_sup_variant_g615
To use these models on the command line or in EPI2ME Labs see below.
Running on command line
To use updated medaka models on the command line simply include the flag --medaka_model with the name of the model.
For high accuracy basecalls:
nextflow run epi2me-labs wf-artic -r v0.3.25 --fastq <PATH_TO_DEMUX_FASTQ_FILES> --medaka_model r1041_e82_400bps_hac_variant_g615
For super high accuracy basecalls:
nextflow run epi2me-labs wf-artic -r v0.3.25 --fastq <PATH_TO_DEMUX_FASTQ_FILES> --medaka_model r1041_e82_400bps_sup_variant_g615
Running in EPI2ME Labs
Update wf-artic
Ensure you have updated wf-artic in EPI2ME Labs and have version >= v0.3.23. Click the workflow and scroll down to find some tools for workflow management.
Click “Update workflow”
If required you can switch the version of the workflow you are using by clicking “Switch revision”
After update the workflow should display the latest version
Run wf-artic with new models
To specify a medaka variant calling model with superior performance on Kit V14/R10.4.1:
Select wf-artic from the workflows menu of EPI2ME Labs and click “Run this workflow”. You will be presented with options for workflow execution. Scroll down to see the “Advanced options”.
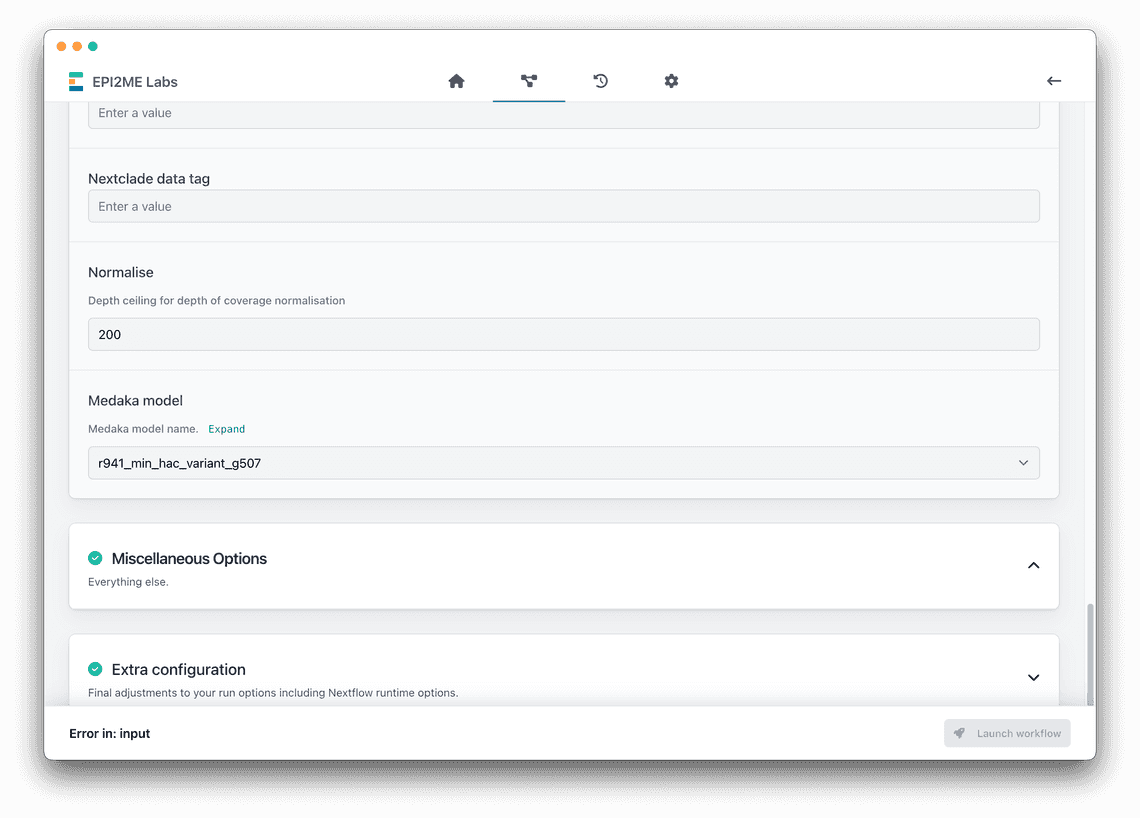
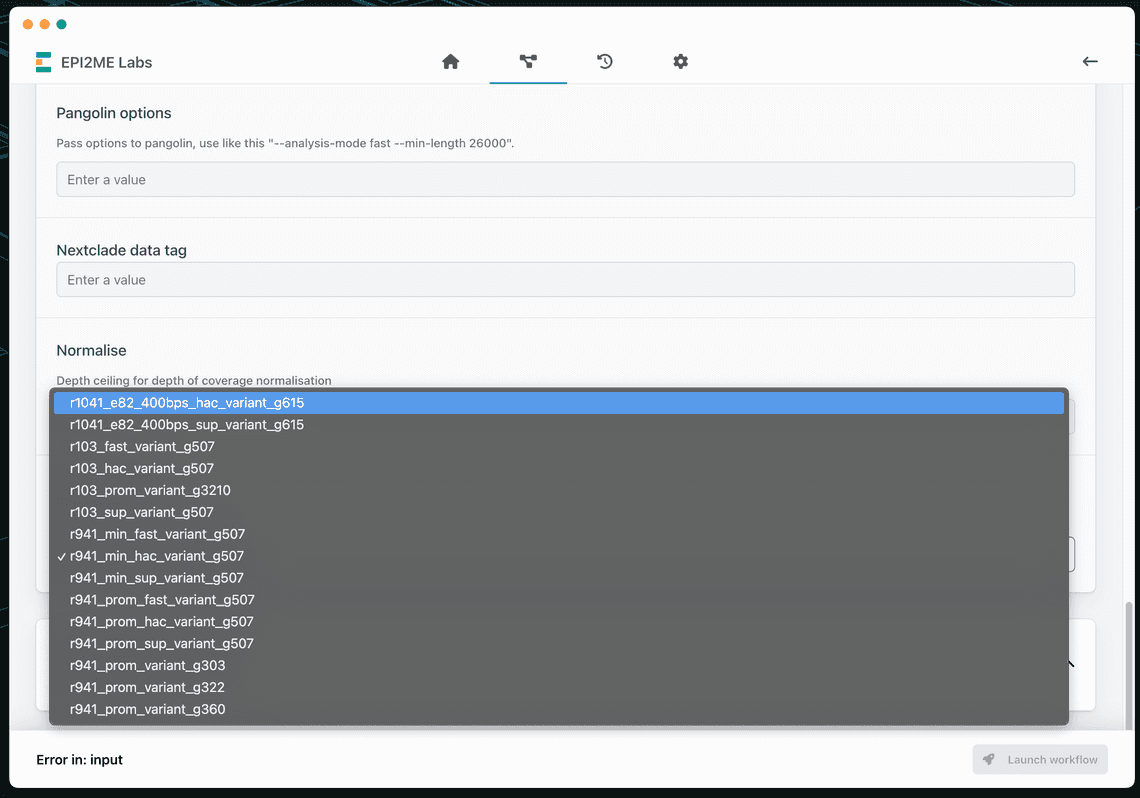
Scroll down to see the medaka model selection drop-down. Select either:
- High accuracy -
r1041_e82_400bps_hac_variant_g615 - Super high accuracy -
r1041_e82_400bps_sup_variant_g615
Share
Table Of Contents
Related Posts
Information

