Two years with Nextflow
The EPI2ME Labs team has now been using Nextflow for almost two years. In that time we have built and released around twenty bioinformatics workflows for Oxford Nanopore Technologies sequencing data analysis. Along the way there’s been a lot of head scratching, bashing of heads on desks, and some swearing. However we’ve also established a set of practices that allow us to rapidly create, build, and deploy workflows within days. Nextflow has become the missing piece in our analysis of DNA and RNA sequencing data.
In this post we will describe our experience of using Nextflow compared to other workflow managers, enumerate some of the issues we’ve encountered, and present our views of how Nextflow ought to evolve to stay relevant.
Introduction
Discussions around workflow managers have a tendency to turn into flame wars akin to classic vim vs. emacs debates. We’ve previously written about why we chose to use Nextflow as our workflow manager of choice. Briefly to recap, and with some confirmation bias from the last two years:
explicit data flow: tasks are stitched together by the developer in a clear manner without relying on implicit matching of filepaths (as with make-like systems),
decoupling of filenames: relatedly, tasks are isolated from each other and communication of data is decoupled from the naming of files,
online analysis: unlike many workflow managers within the informatics space Nextflow is not limited to batch compute on static inputs. It borrows some ideas from modern, continuous ETL systems to provide a reactive system,
excellent support for a multitude of compute engines: to a large extent Nextflow has made distributed compute a solved problem, including support for a variety of software distribution mechanisms.
But this post isn’t about extolling the virtues of one workflow manager or another; more simply stating our experience with using the manager that we have chosen to use. The post was motivated by a Nextflow Language Discussion on GitHub to which we’ve contributed. Here we will elaborate on some of the comments made there and provide further background.
Discussion
The EPI2ME Labs team at Oxford Nanopore Technologies has made an investments in Nextflow, which has paid off through the ease at which we can release new analysis workflows to accompany a line of commercial sequencing products. Our workflows are primarily distributed through Nextflow’s integration with Git repositories, and use conda and Docker to distribute the software used by the workflows. The recently released wf-flu workflow was initially written in a single day from scratch, and could have been released the same day. Our wf-single-cell was ported from a snakemake version created by the Applications Division at Oxford Nanopore Technologies in less than a week. Workflows authored by the EPI2ME Labs team are not only available openly through GitHub but are also installed on our GridION devices, and are available for use through our real-time cloud analysis EPI2ME platform, as well as our EPI2ME Labs Desktop Application.
The power of Nextflow
We’ve been able to achieve this rapid pace of development through the batteries included functionality that Nexflow contains. These are too numerous to list in their entirety (and any who knows me knows I’m mostly a miserable realist; it’s not in my nature to start waxing lyrical about Nextflow here). There are however a few features that we have come to rely on which are worth mentioning.
It’s not obvious from the GitHub repositories serving our workflows the extent to which we leverage some of Nextflow’s integrations and incidental functionality. The first one of note is the way in which Nextflow handles cloud compute resource. For us this is through AWS batch. Within our continuous integration system we run our workflows with sample data on many compute environments including local Linux servers, Apple hardware running macOS, Windows 10 systems, as well as launching workflows into the cloud straight from our test servers with AWS batch. This is a completely trivial affair, we simply provide Nextflow with our access credentials and stuff happens. Nextflow handles moving local data to the cloud, making requests for the required compute resources and downloading results back to us. Admittedly setting up an AWS batch environment itself is not a completely trivial affair, but is within reach of most bioinformaticians or IT system admins.
The second aspect we leverage heavily is Nextflow’s integrations with numerous software deployment environments. All our workflows use conda as a base for software package management. We typically recommend that users make use of the container images created with conda packages and deployed through Dockerhub. These can be used with a variety of container runtimes; in our testing environment all our workflows are tested with at least Docker and Singularity. Again Nextflow manages the use and excution of software in containers for us with no fuss. We were surprised to find just how easy this was with various niceities such as informal script code from the workflow being mounted into the container and available for use within jobs.
Intuition and dataflow
Nextflow has a bit of a reputation for having a steep learning curve to get started. Indeed in the words of one Nextflow developer:
“Like, it’s not hard to learn, but there’s always this sinking feeling that you’re not doing something right, and that feeling never completely goes away.”
- Ben Sherman, Seqera Labs
This comment resonates very much with our experience. Despite the very fulsome documentation and patterns website there have been several occasions where we’ve not been able to understand easily how to implement logic in a workflow that we’ve wanted. We have to admit that some of this difficulty has been our own misconceptions; but it should be commented from experience in helping others that there are aspects of Nextflow that simply aren’t intuitive to many.
This section assumes some knowledge of authoring Nextflow workflows and gets technical fast! Stay with us.
Take the following example, which acts as a model for a fairly common pattern in data analysis. We have a workflow that can process all chromosomes of a human genome independently (Figure 1.). There are several steps however where we can break a per-chromosome task into many smaller tasks computed independently, before the results are gathered together for a single per-chromosome task. Finally the workflow outputs a single result across the whole genome. Traditionally we would think of this as a hierarchy of parallel processes: the workflow forks to produce child processes (one per chromosome), which fork further to produce grand-child processes.
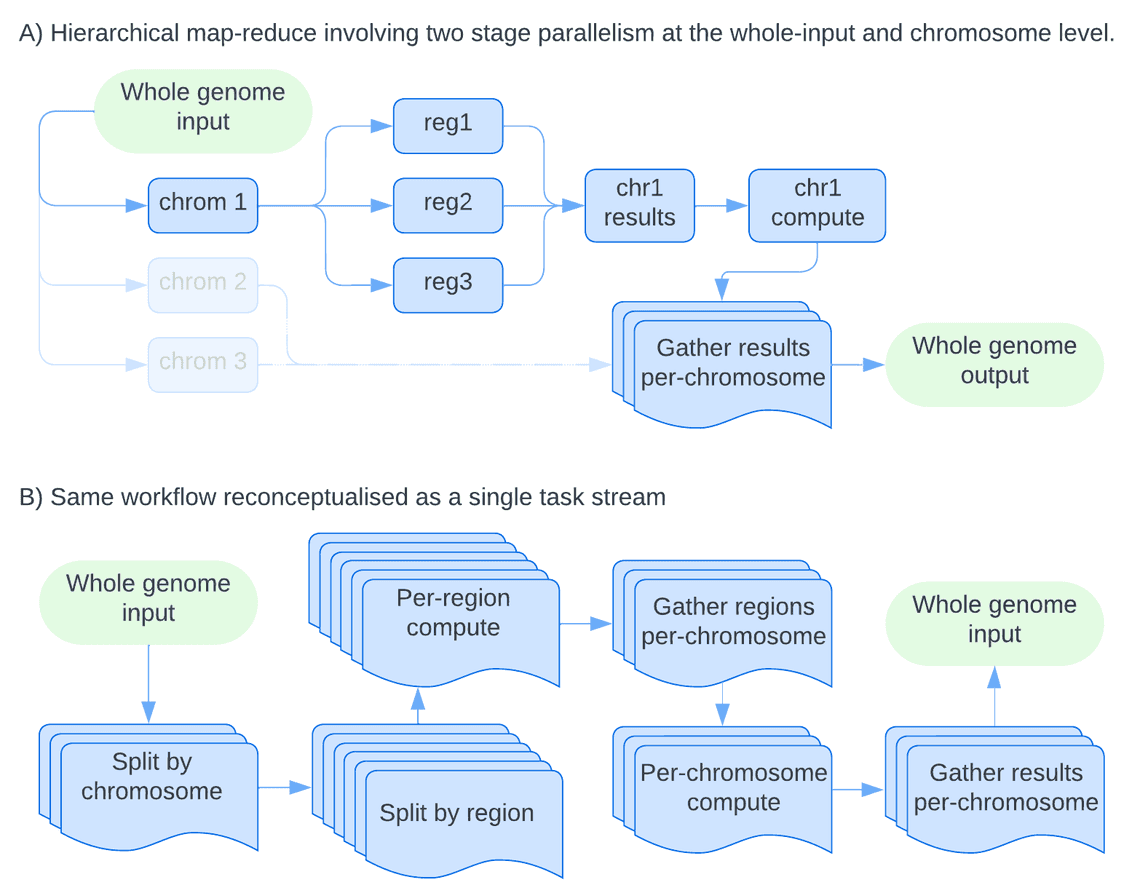
It is tempting to think that such a pattern can be achieved in Nextflow through the use of workflow composition, and it can but not in the way one might expect by mapping workflow scopes to map-reduce operations. Workflow scopes act merely as wrappers around a set of linked processes, for the purposes of the data flow its simply as if the script inside the workflow scope has been cut-and-paste into the main workflow. They cannot be used as a function mapped across a single level of the task hierarchy, i.e. in the current example be used to perform all work for each independent chromosome in an independent manner, with an outer loop over the chromosomes.
To achieve the desired effect in Nextflow we instead decorate the items in our Nextflow data Channels with information for all levels of the task hierarchy, such that their results can be grouped back together later on. A complete example can be found here. The required code turns out to be somewhat simple when you know how: everything hinges on line 129 where the number of sub-chromosomal regions is used to create a key alongside the chromosome, which is used on line 155 in a rather baroque Channel join operation.
To return the original point, the computational pattern used as an example above
is fairly common but needs a bit of insider knowledge to implement. How do we
know that we’re supposed to perform the task like this? We don’t. For instance
there’s only a very small remark in the Nextflow documentation regarding the the
groupKey function of Groovy found in the
groupTuple
section. Having established the correct conceptual model and knowing the
solution somehow hinges on groupKey, how to write the Nextflow code is not
entirely obvious. In trying to implement the pattern we found no fewer than
three historical GitHub issues where users were asking seemingly this question
(or related scenarios) with fragmented responses. We eventually contructed the
solution by piecing together fragments of answers and through trial an error.
It’s interesting to note that number of experienced Nextflow users from the community did not know how to accomplish the task. We start to think why would this be, for such an obviously useful pattern? Our feeling is that there is a lack of more in-depth knowledge in the community around how the internals and scripting layer of Nextflow work and so how anything other that simple linear workflows can be implemented. We’ve experienced a similar phenomenom to that found when dealing with the pandas library in Python: people fumble around with code, copying other snippets until it something works the way they want, and then move on without really understanding why. The cause of this is that various parts of Nextflow are not intuitive to its user base. This is similar to the ideas raised by Ben Sherman that you’re never quite sure whether you’re doing things as intended. The lack of real understanding in the community is something we’ll come back to.
The elephant in the room
Groovy. There, we said it.
Before anyone gets angry, allow us to qualify this observation. For the most part, there is little reason why most people casually writing a Nextflow workflow for their own use need to become experts in Groovy. More important might be understanding rudimentary concepts from functional programming. This is perhaps the key to where many bioinformaticians struggle when starting out with Nextflow. Those with a more biology orientated background and less computer science must grapple with the ideas of higher order functions, iterators and closures whilst they are simultaneously dealing with Nextflow’s idiosyncrasies. When users struggle, they are left with a feeling about what exactly their lack of knowledge is: is it in how they are using Nextflow or their understanding of functionality inherited from Groovy? Where should they go to read up?
We should point out that the Nextflow Scripting section of the documentation serves as a very good primer on the things most users are going to need to know.
A particular point to note is that writing Nextflow script is not synonymous with writing Groovy; that is to say, the Nextflow language is not an extension of Groovy. Some valid Groovy code is not valid Nextflow. Until recently one somewhat confusing difference is that Nextflow functions do not behave as Groovy functions. We first came across this issue, but didn’t realise, a few months into using Nextflow. The issue revolves around the fact that passing arguments to functions in Nextflow does not work the same as in Groovy. Almost the first time we tried to write a Nextflow function we hit the error described in the GitHub issue. As we were new to Nextflow and Groovy we thought it was something that we were doing wrong. Eventually we gave up, bodged the code and moved on with our lives.
Almost a year later we happened to stumble upon the GitHub issue above and realised that other users had had similar issues. After this we were motivated to find a solution. It didn’t take long trawling through the Nextflow source code to realise that the error was indeed in Nextflow and not anything we had been doing. This was somewhat surprising to us: Nextflow is a fairly well used tool at this point, but we had come across a seemingly obvious foible in the language.
To be more constructive in our criticism, it is not that we believe that Nextflow is bad because it is written in Groovy but that it suffers because of it. There has historically been a lack of developers to maintain, support, and extend Nextflow (Figure 2.); it is approximately the product of a single person. This has limited the rate of bug fixes, addition of new features and removal of historical warts for new ideas.
The nf-core project is in some ways another example of this. In addition to providing a set of practices for authoring workflows, the nf-core project adds a suite of tools for maintaining projects and runtime tools adding functionality to the base Nextflow experience. It is notable that these tools are not written in Groovy; it is not the language of choice for even seasoned Nextflow professionals. The existence of the runtime tools begs the question why are such things not simply in the base experience?
The most notable runtime functionality found in nf-core is the creation of command-line help information from a specification of workflow parameters. Yes, it really is the case that Nextflow currently provides no mechanism to validate parameters passed from the user. A developer cannot for example indicate that a certain parameter should be an integer between 1 and 10. It does however try to perform automatic coercion of parameters to types, with sometimes frustrating consequences.
So should Nextflow be rewritten in a less esoteric language? Probably not, but we do think many of rough edges and questionable design choices in the language would have been smoothed off by now if it were written in a language that promoted a higher level of engagement with developers. Unfortunately we do not believe there is an easy solution to this problem: Groovy is not a commonly known language either within Nextflow’s audience or the wider data analysis and computing community.
Growing pains
With the above considered we wonder if Nextflow is in someway a victim of its own success. We have encountered various examples of issues we’d have expected to have not occurred or to have better solutions by this point in Nextflow’s life. Afterall, we are coming to Nextflow somewhat late in the game. Is it that there are not a sufficient number of active developers to keep pace with user requests for new functionality and the fixing of issues? Certainly examining contributions from the community, other that contributions to the documentation, show many add extensions of a discrete type such as adding the ability to run jobs on a new type of job scheduler. Many do not affect the codebase in a more extensive manner.
Examining issues reported through GitHub does not provide conclusive evidence of
this hypothesis, Figure 3. We searched GitHub for the author of all issues on
both the nextflow-io/nextflow project and all projects under the nf-core
organisation. We included the latter as we presumed that users may have started
to ask questions through nf-core that they would have otherwise ask through
nextflow. As it transpires, nf-core contributes a large part to the total
community: we find at most a 10% overlap between the two groups of issues for
any one half-year.
We do however admit that perhaps early 2018 serves as a turning point in at least a figurative sense. The number of users at this time became more than one user per three days reporting at least one issue. That is not an insignificant number of users, especially considering the very few developers (even fewer working full-time on the project) revealed by of analysis of contributors (Figure 2.).
Final words
We would be remiss to not mention what we believe is the biggest issue currently facing Nextflow as a language. This is something already mentioned on the Nextflow Language Discussion. Our biggest pain point with Nextflow is the debugging experience when writing Nextflow scripts. It is not uncommon to see errors like the following:
Script compilation error- file : workflow.nf- cause: Unexpected input: '{' @ line 19, column 10.workflow {^1 error
What does this mean? Simply that we have an error somewhere in our workflow script: we get no more help! This leads to a very frustrating debugging experience, especially when taken in context with the other issues raised in this post.
Summary
In this post we’ve explored our experience of using Nextflow over the last two years. We’ve spent a lot of time highlighting some of the larger difficulties we’ve experienced and tried to rationalise why these may have occured in terms of the number of active users and developer using and working with the software. Some of the issues we’ve had have been conceptual in nature, heightened by a lack of knowledge and support in the community. Others were rather more surprising considering the age and maturity of Nextflow – historical warts and rough edges exist of which users need to be aware.
So what are we to do? The first is that we encourage Nextflow users to submit issues on GitHub, and contribute to the Nextflow language improvement discussion. For those who are able (and those who wish to learn), we urge you to get involved with Nextflow and expand community contributions. We will certainly keep using Nextflow and continue to break it!
Nextflow provides us and our users with many benefits portable execution on many compute platforms to ease of distribution and explicit data flow and automatic implicit parallel compute (when you know how). We look forward to a time when with a better debugging experience, the parameter system is rationalised, and the language simplified and cleaned up. Who knows, might we one day soon have a DSL3?
Share
Table Of Contents
Related Posts
Information

