Simulating sequencing datasets

Bioinformatics tools and pipelines, as with any analysis methods, need to be tested to ensure they work as expected. This requires benchmarking them to determine their accuracy and to assess the impact of various factors on the results. Such a task requires data for which the ground truth is known, i.e. knowing what a 100% accurate result should look like. While there are some situations where users may be able to rely on real sequencing data from robustly characterised mock samples in the lab, simulating datasets on a computer is commonly the only practical and reliable option.
But how do you go about simulating datasets fit for this purpose? This is a huge field of research, as simulating realistic sequencing data is not trivial. The goal is to create a set of sequencing reads that reflect not only the biology of an underlying hypothetical sample, but also the patterns and noise associated with real sequencing data. Such variation results from a variety of sources, including:
- library preparation - preparation introduces several sources of variation. Fragmentation of molecules, errors and biases introduced by PCR amplification, formation of chimeric molecules, and biases introduced through ligation or hybridization of molecules to artificial constructs or probes, all contribute to the complexity of the resulting dataset.
- sequencing - the sequencing process itself, including the sequencing platform and basecaller, can introduce both systematic and random error into the recorded base sequences.
Having unrealistic data can prevent you from getting accurate benchmarking results, and so your choice of methods for simulating data needs to be considered carefully. But don’t let this put you off! This guide will help you to understand the challenges associated with simulating sequencing datasets, so that you can avoid potential pitfalls and gain reliable benchmarking results.
Numerous genome simulation tools exist to automate many of the steps involved in simulating sequencing datasets, with new ones frequently becoming available and each tailored to different scenarios.
While we won’t delve into the full range of simulation tools here (due to their sheer abundance and variety), we’ll cover the general approaches employed by the different methods and mention a few examples towards the end.
General approaches
The most generic, or at least brute force, approach to simulating sequencing data would be start from known reference sequences and fully replicate biological and experimental variation. Such a method gives precise knowledge and traceability of all the reads that come out of the simulation. However this method naturally requires exquisite knowledge of physical processes in the laboratory and the sequencing system, so can be hard to accomplish.
An alternative approach is to manipulate real sequencing reads in a controlled fashion. This has the benefit of not needing to characterise explicitely the complete experimental setup, but does require having adequate knowledge of precisely what was sequenced in the first instance.
We discuss both methods below.
Simulation from reference sequences
Simulating sequencing reads starting from a small collection of reference sequences can be described as a “generative approach”. It aims to create simulated reads by applying a chain of (probabilistic) logical transforms which aim to reflect reality.
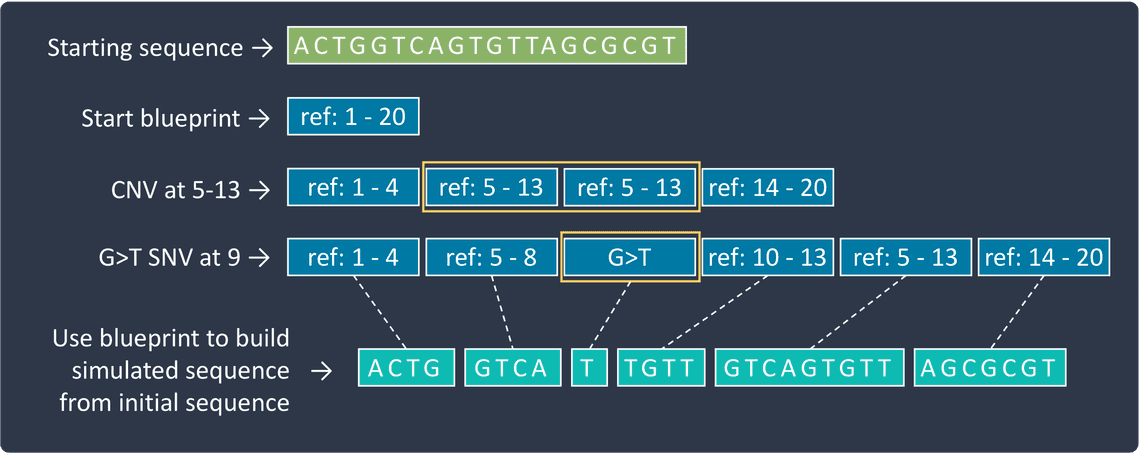
The first step is to simulate biological variation that might be present in a sample, that is not present in our collection of reference sequences. We can introduce mutations in the reference sequences, that are then used in subsequent steps. For example, we could take FASTA file containing a genome sequence and swap the base at position 100 from a T to a G to simulate a point mutation. Or we could copy the sequence from position 50-59 and insert it between bases 59 and 60, to simulate a copy number variant at position 50-59 with a copy number of 2.
One of the challenges with modifying a genome sequence is keeping track of where each base corresponds to in the original sequence. As soon as any insertions or deletions occur in a sequence, the Nth base in the original sequence may not match up with the Nth base in the modified sequence, and so consideration needs to be given into working out where any subsequent variants should be placed. Various approaches are used to handle this. One option is to incorporate variants in order from last to first so that the sequence prior to the last incorporated variant position always corresponds to the original sequence. This works well when the larger insertions don’t overlap other variants, but in situations where they do, more sophisticated methods are required. Another option is to use a “blueprint” representation of the sequence, that can more easily be manipulated during the simulation. This blueprint takes the form of a list of sections, each representing either a portion of the original sequence, in which case only a start and end position is recorded, or instead a variant sequence. The list of sections is sequentially modified to represent each new variant, and once all the variants are incorporated, it can be used as instructions for building the final simulated sequence. For each section either the relevant portion of the original sequence or, if present, the recorded variant sequence is copied and concatenated together to form the full simulated sequence.
Once we have our modified reference sequence, we next need to generate reads from it. The aim to mimic real library preparation and the sequencing process by essentially selecting regions from the simulated genome sequences, using specified location and length distributions. However, achieving this in a realistic manor is a significant challenge. There are at least two important considerations, elucidated below.
Read coverage
Almost all library preparation and sequencing methods result in at least some degree of non-uniform read coverage along a genome.
Further biases may also manifest in the direction of reads, with a tendency for more reads to originate from either the forward or reverse strand.
In short-read sequencing, GC composition is a strong factor in how efficiently a region is sequenced, with many short read simulators taking this into account.
Further consideration needs to be taken when simulating amplicon, whole-exome, or other targeted sequencing datasets.
While simply limiting the reference sequence sampling to the relevant regions of the genome may be sufficient in some experiments being performed with simulated data, more tailored approaches are needed to reflect the nuances of these datasets.
Incorporating sequencing error
Real-world sequencing data contains various errors; if it didn’t then variant calling would be a lot easier!
These errors need to be incorporated into the simulated reads, with base quality scores that reflect the uncertainty.
At a basic level this might, for example, involve measuring empirical errors and quality distributions and applying these to the “perfect” reads derived from the previous step.
Such models can be made increasingly sophisticated and incorporate models of the surrounding sequence and read position context when simulating quality scores and errors.
For example in short-read sequencing by synthesis, a T nucleotide following GG is more likely to be incorrectly sequenced than a T following AA (Stoler and Nekrutenko 2021).
Similarly positions closer to the ends of reads from such sequencing methods can also have an increased error likelihood.
Taking the idea of the generative approach to the extreme one might instead physically model the measurement process and apply the same data processing as would ordinarily be applied to real data. Such an approach was previously used within scrappie, where a MinION “squiggles” (ionic current traces) could be simulated from a provided nucleotide sequenced, additional noise and variation added, with the resulting signal passed to a basecaller to produce a simulated basecall.
Many such simulation methods allow the user to train their own models on real data in order to account for the specific chemistry, library preparation, and genome context relevant to their study. Even so, the reads generated by these methods can vary in how realistically they reflect real data.
The complexity in accurately modelling physical processes is the main rationale for using alternative simulation approaches which utilise and modify real sequencing datasets.
Simulation from real-data
As a somewhat simpler approach, an alternative strategy for simulating read sets is to sample reads from existing experimental data. This does not require any in-depth specialist knowlege and so can more easily achieve the desired datasets.
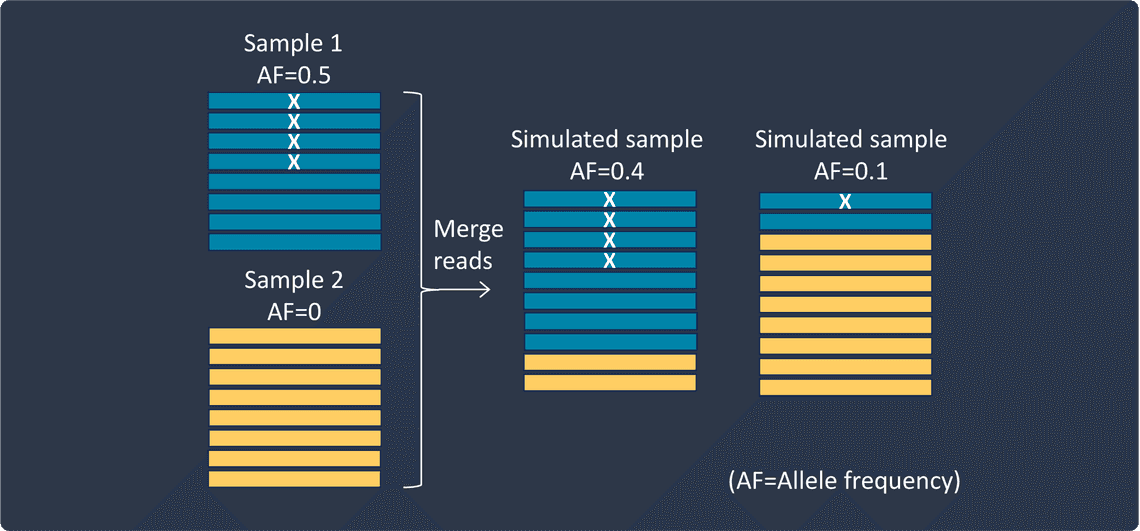
The basic idea is to subsample reads from an existing biological sample and merge them together with reads from other samples at required frequencies. This can generate datasets with a specified coverage, and with variants, species, or lineages at specified frequencies.
The process isn’t without its disadvantages however. Firstly flexibility is limited. The simulation is restricted by the coverage, variants, species that are available in existing datasets.
Secondly, the technique requires thorough characterisation of the initial datasets with respect to the type of benchmarking planned. If you want to test variant calling at different allele frequencies, then you need to know the positions of all true variants in the initial datasets, and if you want to test lineage analysis, then you need to be confident of the lineage composition of the initial datasets. If this characterisation is not accurate, then any benchmarking performed on samples created from them will be unreliable.
In order to combat the first of these disadvantages, a modified method is to introduce mutations into the library of sequenced reads. This approach avoids many of the issues associated with artificially generating reads de novo, though it comes with some difficulties of its own.
The process relies on having accurate initial identification of the reads which are mutated: if we cannot confidently identify the reads then we will be introducing mutations other than those which we thought we were creating. The identity of reads is typically made through alignment, and introduction of variants is through known genomic co-ordinates. The mis-indentification (and therefore application of variants) can therefore occur due to ambiguities in the read alignment process.
The ambiguous alignment of reads is a whole topic of conversation in itself; let us discuss briefly consider one such example. Consider the case where a genome contains non-identical repeats. When aligning reads to this genome it can be the case that there is an ambiguity as to which repeat the read is derived. There can be errors and biases in this assignment; particularly if sequencing errors conflate with the differences between the repeats. If we apply variants to all reads which align to one copy of a repetitive element, we may in fact be applying variants to reads whose true identity is a second copy of the element.
Having introduced variants, this misalignment effect can work the other way round. The modified reads may be significantly different from the original seed read, and may now more properly align to another locus of the genome entirely; however our synthetic dataset has the read aligned at the original location. In a real sequencing dataset with the variant present, a read may align to the alternative location: our simulated read set does not now reflect a real set od aligned reads. If we were to now benchmark a variant caller with the synthetic set we may obtain better results than with real reads (natively containing the variants). Realigning the synthetic reads following artificial introduction of variants would better model a real experiment, and the resulting inaccuracies to be accounted for in subsequent benchmarking.
Finally, care should be taken when incorporating variants to ensure that other properties of the data are maintained. For example: base qualities. Suppose we insert two Gs before an existing pair of Gs. In real data this would often result in low quality scores for the whole stretch of Gs due to it creating a 4-base homopolymer. Similarly if a GGT trimer has an A inserted before the T, then in real data this should result in an increased quality score for the T.
How to get hold of well characterised real datasets
So those are two different approaches used to simulate sequencing datasets. You might now be asking, “how do I get hold of some thoroughly characterised real datasets?”
The best way to get a well characterised dataset again depends on your specific use case. Previously characterised datasets are often available for common scenarios so it’s worth searching for one, or having a look at what’s been used in other simulation studies in your field. For human data, consortia exist that provide extensively characterised samples, such as the Genome in a Bottle Consortium. Among other methods, the GIAB consortia employ son/father/mother trios to determine sets of high confidence variant calls. Other options may be to generate data yourself in such a way that you are confident of the ground truths. For example, sequencing pure isolates of species for use in simulating datasets for abundance testing.
Which approach do I use?
I need to test how my pipeline performs with lower coverage
Simple! Use the second approach by subsampling to reduce coverage of an existing dataset, either from FASTQ or BAM files:
samtools view -bh -s {DOWNSAMPLE_FRACTION} {INPUT BAM} > {SUBSAMPLED_BAM}
Okay, that was a fairly obvious one to start with, but it does get more interesting!
I need to test my metagenomics abundance estimation pipeline
The best option for this would likely be the second approach above, providing you have real datasets available for a sufficient variety of species, and importantly, of sufficient purity. In metagenomic datasets we would hope that the relative abundance of reads should reflect the abundance of species in a sample. However in reality, factors such as GC content and extraction and library preparation biases result in non-uniform sampling of genomes. This should therefore be reflected in the simulated datasets. You may also want to add in some reads from unknown microbes or from human contamination, to add to the authenticity of the simulated samples.

If the above approach is not feasible due to lack of datasets, then another option is to throw in a bit of the first simulation approach!
I need to test how my pipeline performs with lower allele frequency
Use the second approach to mix reads from two different samples at various proportions, resulting in variants with altered allele frequencies.
I need to test my short variant caller on a specific set of novel variants
This requires simulating new variants through either of the approaches. As we mentioned before, these each have their own drawbacks so it’s worth considering which suits your needs best before deciding on one.
When using the first approach you can simulate both the reference sequence and novel variants in proportions corresponding to any desired allele frequencies for the study.
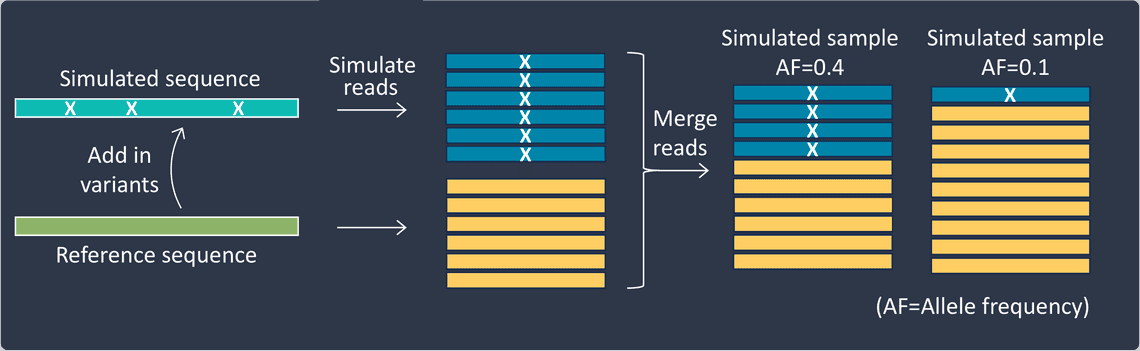
For the second approach, you’ll need to acquire real sequencing dataset for which all true variant positions are reliably known (such that these true variants don’t conflate the study). You can then swap bases at the required positions in a portion of reads covering a simulated variant position.
I need to test the performance of my breakpoint-based copy number variant caller
Breakpoints and complex structural rearrangements cannot be modelled easily with the second approach. It is possible in theory to do so by cutting and joining reads together, but this quickly becomes a headache in practice! That leaves us with the first approach probably being our best option. Copy number variants or translocations can be added into a genome sequence and processed to create reads that represent those structural rearrangements.
I need to test my tumour evolution analysis pipeline
We’ve come to our last scenario! Tumours are constantly evolving, with sub-populations of cells forming that contain distinct sets of genetic mutations. Tumour evolution analysis covers a range of techniques that can be applied to tumour sequencing datasets, and which aim to characterise the distinct cell populations and uncover the evolutionary relationships between them.
But how to simulate suitable benchmarking datasets for them?
This requires generating datasets that represent a phylogenetic tree of evolving cell populations, all descended from the tumour’s cell of origin. Simulating such a dataset can be achieved by using either of the approaches, and extending many of the above scenarios in such a way that once one cell population is simulated, subsequent populations are then simulated by acquiring all variants from their parent cell population and incorporating additional new variants.
You’ll probably want to first simulate a germline “cell population”, followed by a cell of origin “cell population”. After that, divergence of cell populations can be achieved by adding distinct sets of variants to two populations originating from the same parent, and shorter or larger evolutionary distances can be reflected by altering the number of new variants added.
Once the reads for each cell population are created, they can be merged at required frequencies. Different samples can be created that contain varying proportions of each cell population to reflect different tumour sampling regions or time points through therapy.
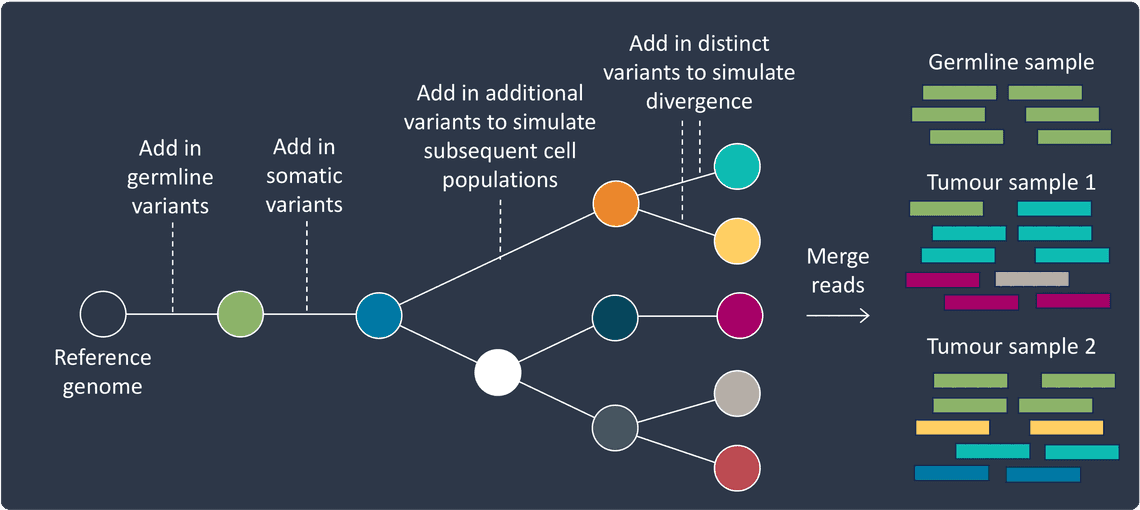
It is not necessary to merge, or even generate reads, from every stage of the phylogenetic tree. In tumours, cell populations can die off from being outcompeted by others or as a result of treatments, or they might only be present in certain regions of the tumour.
A similar approach to simulating tumours might be useful for simulating microbial evolution.
One final point to mention about this scenario: an important feature of tumour evolution is that the genome can become very messy and complex. For example, copy number variants can overlap other copy number variants, and short variants can occur on one or multiple copies of a multiplied region depending on whether the short variant occurred in time before or after the copy number variant. These features can make it even more challenging to analyse tumour evolution. So if the analysis you’re wanting to benchmark could be affected by this complexity then it’s important to include them in your simulated datasets.
Simulation tool examples
You’re hopefully now aware of how to simulate sequencing datasets in various different scenarios. Many more applications will exist and by understanding the considerations needed for the above, you will have an idea of how to decide on the best approach for others.
- Icarust - a complete MinKNOW simulator including squiggle and basecall simulation, Munro et al. 2024
- NanoSIM - Simulates nanopore long-read sequencing data, Yang et al. 2017
- DeepSimulator1.5 - Simulates nanopore long-read sequencing data, via simulating the raw electrical signals Wang et al. 2020
- A recent review of other short-read simulators - Milhaven and Pfeifer 2023
- CAMISIM - Metagenomics simulator, incorporating various other short and long-read simulators, Fritz 2019 et al.
- HeteroGenesis - Simulates genome sequences for complex somatic samples, Tanner et al. 2019
What next?
For a guide on how to perform benchmarking once you have your simulated datasets, see our recent blog post on Benchmarking performance with sensitivity, specificity, precision and recall.
Happy simulating!
Share
Georgette Tanner
Clinical Bioinformatics Software Developer
Related Posts
Information
