Comparing sequences with dot plots

If you had to make a list of the most valuable and insightful ways of visualising sequence data, would the humble but powerful dot plot be on that list? Dot plots are a popular tool with which you might already be familiar. We use them in several of our workflows. In this blog post we’ll look at when to use dot plots, and how to create and interpret them.
A dot plot is one way of comparing two sequences to find similarities and differences between them. It can be one sequence with itself (self comparison), or comparing two different sequences (cross comparison). A dot plot can be used for comparing any type of sequence including DNA, RNA, and amino acids. They allow you to quickly identify regions of interest in a sequence that may require further investigation. Other use cases of dot plots include:
- Obtaining a global perspective of differences between two sequences.
- Detection of repetitive elements such as tandem repeats or transposons within one sequence.
- Finding homologous regions between two sequences.
- Quality control of de-novo sequence assembly to assess continuity and completeness of assembled sequences.
- Visualising mutations such as insertions and deletions between a reference and sample sequence, as well as inversions, translocations and more complex rearrangements.
For instance, we use dot plots in the context of plasmid sequencing (with wf-clone-validation) to perform a self comparison of the de-novo assembled plasmid sequence to help identify expected or unexpected repetitive elements.
Background
To create a dot plot, we need to associate each of the two sequences with an axis of a 2D scatter plot. The plot depicts regions of the two sequences which are similar, often referred to as homologous regions. Marking a point on the scatter plot for every base pair that is the same between two sequences would lead to a very noisy dot plot. For example, it would be silly to mark a dot at every possible co-ordinate where the two sequences have an ‘A’. Instead similarity is only considered worthy of being marked when it occurs over longer distances. We mark only subsequence matches of a predetermined length. Note that the definition of “matches” in the context of sequence alignment may include approximate matches; some variation between two subsequences of the two sequences is allowed.
So we see that when creating dot plots, it is useful to understand a little about the parameters involved in sequence alignment. The extent to which short subsequences of two longer sequences are similar, but not necessarily identical, is often parameterised by three parameters. Alignments are “scored” according to the so-called match, mismatch, and gap penalties:
- The match score is positive and awarded for identical base matches between a sequence.
- The mismatch score subtracts from the overall score as a penalty for base differences.
- The gap penalty also subtracts and is given when one sequence has a missing base relative to the other sequence.
More elaborate scoring schemes exist (particularly for protein alignments), but it suffices to say that subsequence alignments can be scored to provide a quantitative measure of their similarity.
In order to create a dot plot, we must decide which subsequence alignments are similar enough to be plotted. A simple and common method is to filter alignments by their length and their score. Decreasing the subsequence length or alignment score can reduce stringency to reveal more subtle structural relationships, but can also increase background noise. Raising the stringency of the filtering will produce cleaner plots at the expense of missing information.
Various online dot plot tools may implement different scoring systems and thresholds and therefore produce qualitatively different looking plots.
Creating plots
We’ve discussed a little about the background theory of how data underlying a dot plot can be created. How though do we go about creating one? Many tools and packages are available for sequence alignment. Several such tools exist with special considerations for creating dot plots, such as:
- Vectorbuilder dot plot is a simple online tool for comparing smaller sequences.
- The Genome Evolutions Syn Map online programme is great for exploring homology between larger sequences. It can retrieve the FASTA sequences required and create a dot plot which you can use to select regions of interest to inspect more closely.
- Mummer4 is a great offline option which has a nice tutorial.
- Minidot is an R package for visualising dot plots of minimap mappings.
In our clone validation workflow we use the Last tool to create self alignments and plot the outputs using Bokeh.
We first create an alignment database using the lastdb command-line tool, and then proceed to perform alignments using the lastal command.
Using, as an example, the DNA sequence of the MYC associated zinc finger protein (MAZ) we can run:
lastdb db.lastdb maz.fasta
to create an alignment database.
The database provides data structures which allow computationally efficient sequence similarity searches to be made.
We then use the lastal command with a query sequence.
As this is a self comparison we use the same FASTA file again.
lastal -m 1000 -w 10 db.lastdb maz.fasta > maz.maf
This outputs a MAF file which includes a list of the alignments found.
We have increased the maximum initial matches per query position (-m) parameter from a default of 10 to 1000, to increase sensitivity which for this short sequence comes at a minimal computational cost.
We also reduced the offset distance for suppressing repeats inside exact matches (-w) from a default of 1000 to 10, again increasing sensitivity for this small sequence allowing us to visualise the self comparison in finer detail.
Then we used the following python function to create the Bokeh plot from the MAF file.
# Import bokeh plotting librariesfrom bokeh.models import ColumnDataSource, Segmentfrom bokeh.plotting import figureimport pandas as pddef dot_plot_assembly(maf_assembly):"""dot plot of assembly using a .maf format file."""# Create a bokeh plotplot = figure(width=400, height=400,min_border=0, x_axis_label="First sequence position",y_axis_label="Second sequence position", title="Dot plot")plot.toolbar_location = None# Iterate through maf file to create a dataframerecords = list()with open(maf_assembly) as maf:while True:line = maf.readline()if line.startswith('#'):# get read length available in the MAF generated with LASTif 'letters' in line:read_length = int(line.split('letters=')[1])continueelse:continue# a is for each alignmentelif line.startswith('a'):# take successive 's' linesr1 = maf.readline().split()[1:5]r2 = maf.readline().split()[1:5]maf.readline()records.append(r1 + r2)elif line == "":breakelse:raise IOError("Cannot read alignment file")# take reference start, length and orientation# and query start, length and orientationnames = ['ref', 'rstart', 'rlen', 'rorient','query', 'qstart', 'qlen', 'qorient']df_all = pd.DataFrame(records, columns=names)df_all = df_all.astype({'qstart': int, 'qlen': int, 'rstart': int, 'rlen': int})# If query orientation is +df = df_all[df_all.qorient.isin(['+'])]# create query and ref end columns by adding length to startdf['qend'] = df['qstart'] + df['qlen']df.loc[df['rorient'] == '+', 'rend'] = df['rstart'] + df['rlen']# If reference orientation is negative switch reference start and enddf.loc[df['rorient'] == '-', 'rend'] = df['rstart']df.loc[df['rorient'] == '-', 'rstart'] = df['rstart'] - df['rlen']# Add forward lines to plotsource = ColumnDataSource(df)glyph = Segment(x0='rstart', y0='qstart', x1='rend', y1='qend', line_color="black")plot.add_glyph(source, glyph)# if query orientation is -df = df_all[df_all.qorient.isin(['-'])]# If the orientation is "-", start coordinate is in the reverse strand (maf docs)# Therefore as plot will be + vs + query start needs to be flippeddf['qstart'] = read_length - df['qstart']df['qend'] = df['qstart'] - df['qlen']df['rend'] = df['rstart'] + df['rlen']# Add reverse complement lines to plotsource = ColumnDataSource(df)glyph = Segment(x0='rstart', y0='qstart', x1='rend', y1='qend', line_color="red")plot.add_glyph(source, glyph)return plot
This creates the following plot which as you can see consists of red and black diagonal lines in varying orientations. The x-axis is showing the base pairs of the (first) reference sequence and the y-axis is the base pairs of the (second) query sequence. So what does all this mean? It may look complicated at first, but read on to find out more.
Interpreting dot plots
In the previous section we discussed how to create dot plots by aligning sequences using lastal and plotting its outputs using Python.
In this section we will discuss what can be discerned from such plots.
Self comparison of a sequence
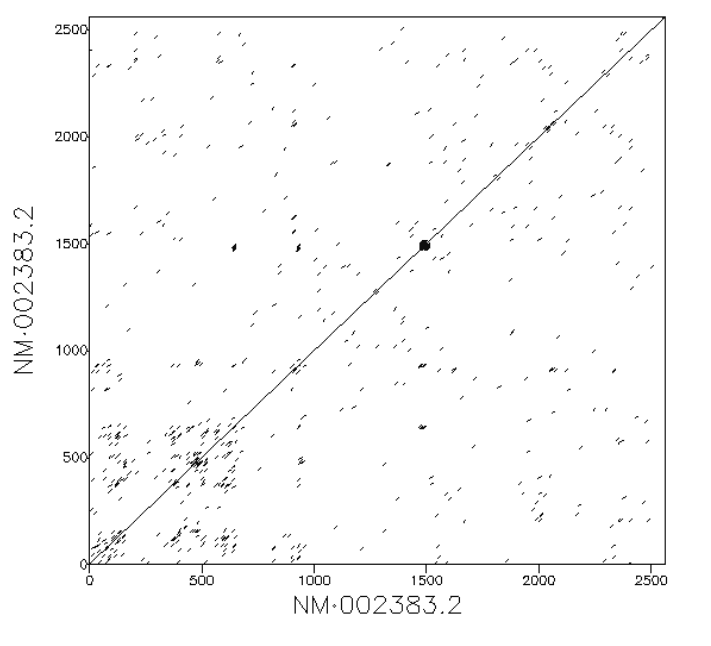
If two sequences are exactly the same we expect to see a diagonal line. This represents the fact that the every position in the first sequence matches exactly the equivalent position in the second.
A sequence showing a duplication
If there are duplications within the sequence we would see parallel diagonal lines. In the example below the 10 kb input sequence consists of a duplicated element of 5 kb length. We see three parallel lines. The central diagonal line along y=x depicts regions aligning to themselves. The lines above and below the diagnonal indicate distinct homologous copies of sequence aligning to each other. The upper line is the second copy of the repeat in the reference sequence aligning to the first copy in the query sequence. The lower diagonal is the first copy in the reference sequence aligning to the second copy in the query sequence. If there were five lines this would indicate three duplicate copies, seven lines would indicate three copies, and so on.
A Highly Repetitive sequence
In an extreme case of the above, highly repetitive regions will appear as many small parallel lines which resemble a square:
Insertions and deletions between two sequences
Insertion and deletion are terms used to denote additional or missing stretches in one sequence with respect to another. Subsequences in the second sequence that are not present in the first sequence are termed insertions. These additional sequences have no match in the first sequence; they cause a break in the main diagonal line and a jump up slightly. Deletions are the corollary of this: they are missing subsequence in the second sequence relative to the first. (Alternatively they can be described as an insertion in the first sequence relative to the second). A deletion appears in a dot plot as a jump sideways in the main diagonal line.
Inversions
Inversions are subsequences which occur in the opposite direction in one sequence relative to another. Inversions and inverted repeats appear in dot plots as lines perpendicular to, and intersecting, the main diagonal. Many tools for creating dot plots will colour these lines differently to help distinguish when they are overlapping with forward repeats. In our workflows inversions are coloured red.
Summary
The assemblies and resulting dot plots in the clone validation workflow are usually small and simple. However, you can also create dot plots for large scale genomes with multiple contigs, which can reveal interesting patterns. Sometimes you will need to tweak available parameters to fine tune the sensitivity to make the plot meaningful.
We hope this post showed how useful dot plots are for comparing two sequences and how easy they are to interpret once you recognise the common patterns. Why not have a go at creating your own with some sequences of interest using one of the offline or online tools listed above?

