SARS-CoV-2 Midnight Scheme Update
Introduction
This blog post will explain the changes we are making to the primer scheme naming in wf-artic due to underlying changes in the ONT Midnight SARS-CoV-2 sequencing kit. We will also describe a new feature of wf-artic allowing specification of custom primer schemes.
SARS-CoV-2 Genome Sequencing by Tiled Amplicon
Tiled amplicon sequencing allows us to cover small genomes with short slightly overlapping PCR amplicons. This approach has a number of advantages, possibly the most important being that we can still recover full or nearly full genomes from samples with a low viral load or degraded RNA.
To achieve this tiled amplicon sequencing approach, pools of primer pairs are used to create the amplicons that cover the whole of the genome. For SARS-CoV-2 we analyse this data using the Network Artic FieldBioinformatics tool using wf-artic. This tool requires a bed file of the primer locations used in the assay. These primer “schemes” provide the start and end position of each primer but do not include the primer sequences as these are not required by the workflow.
The primer scheme bed files are required for a couple of reasons:
- Determining which amplicon a read has been generated from
- Trimming of primer sequences from the ends of reads to prevent issues during data analysis
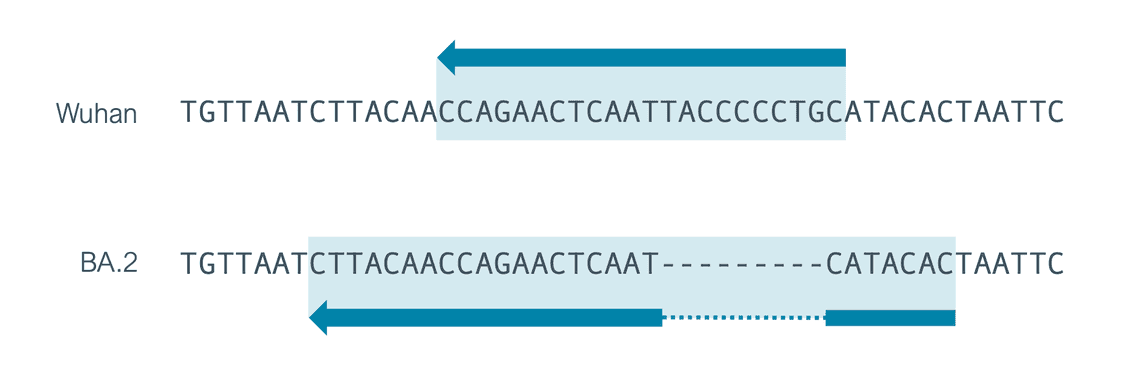
Previously the wf-artic workflow included a single Midnight primer scheme called V1200. Previous additions of primers to ONT Midnight have been sequence only changes and have therefore required no changes to the primer scheme. Version 3 of the ONT Midnight kit adds a new primer to tackle the drop out of amplicon 21 as a result of a 9bp deletion in this region in BA.2 samples. This requires a new scheme. We are therefore, after consulting members of the community, renaming our Midnight schemes to be in line with our physical kit versions.
New schemes
We are making it clearer in the wf-artic repository which scheme should be used with each physical assay in addition to making versioning for Midnight schemes more transparent.
The schemes now included in wf-artic are:
- ARTIC: Original Network Artic primer scheme of 400bp amplicons (Network ARTIC & Josh Quick & Nick Loman)
- Midnight-IDT: This represents the assay produced by IDT and designed by the community (Nikki Freed & Olin Salander) including the recent additions and changes to deal with new variants.
- Midnight-ONT: This represents the assay originally designed by the community (Nikki Freed & Olin Salander) and produced by ONT, but with primer additions and improvements to deal with recent variants of SARS-CoV-2 that differ from those contained in Midnight-IDT.
- NEB-VarSkip: No changes here other than a clearer folder structure.
So when running ARTIC or wf-artic we specify the version as <ASSAY>/<VERSION> where assay could be, for example Midnight-ONT and the version could be V2, this would be written as Midnight-ONT/V2
The rule of thumb for Midnight kits bought from ONT is that the kit version will always pair with the scheme version e.g. EXP-MRT001.30 should be paired with primer scheme Midnight-ONT/V3. If you use the V2 Midnight primers from IDT then you would use Midnight-IDT/V2 as the scheme name.
ONT Midnight kit version 3 primer additions
The table shows the additional (i.e. original V1&V2 primers remain in the mix) primers added to Midnight-ONT kit V3 to address BA.2 amplicon 21 drop-out and provide general performance improvements.
| Primer | Start V1&V2 | End V1&V2 | Sequence V1&V2 | Start V3 | End V3 | Sequence V3 |
|---|---|---|---|---|---|---|
| 21R | 21621 | 21642 | CCAGAACTCAATTACCCCCTGC | 21615 | 21645 | CTTACAACCAGAACTCAATCATACAC |
| 22R | 22591 | 22612 | CGCCACCAGATTTGCATCTGTT | 22591 | 22612 | CGCCACCAaATTTGCATCTGTT |
| 23L | 22512 | 22537 | ACTTTAGAGTCCAACCAACAGAATCT | 22512 | 22537 | ACTTTAGAGTtCAACCAACAGAATCT |
| 24L | 23519 | 23544 | GCTGAACATGTCAACAACTCATATGA | 23519 | 23544 | GCTGAAtATGTCAACAACTCATATGA |
While the additional 22R, 23L and 24L primers represent single base changes (lowercase), 21R has been redesigned to deal with a 9bp deletion found in the primer region in BA.2.
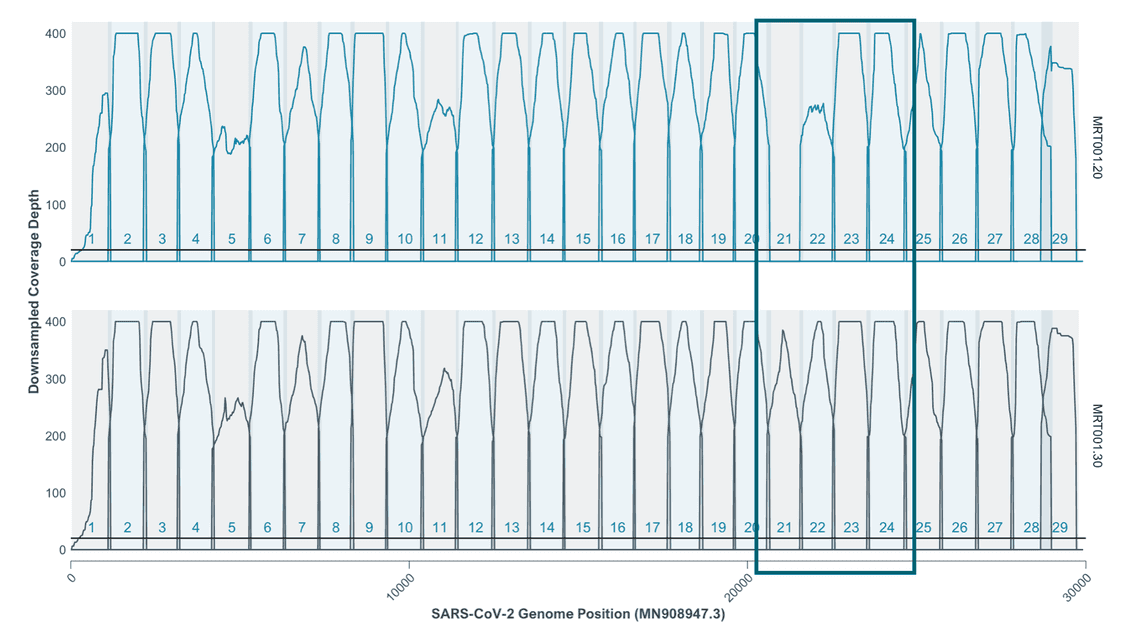
The improvements are clear on a BA.2 sample of ct 20 sequenced using Midnight V2 and Midnight V3:
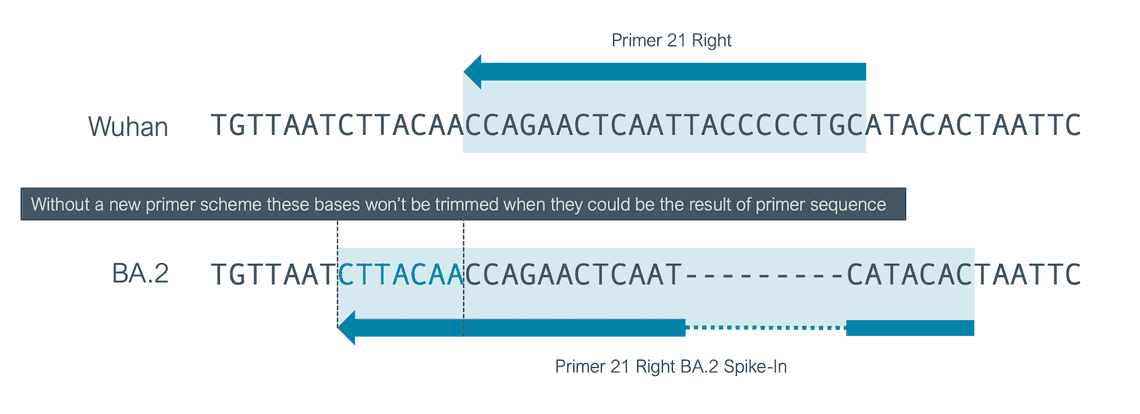
To account for the addition of a primer 21R with a different position we decided to use a scheme containing a single entry for 21R but expand this primer region to cover the whole of the additional new 21R and the original 21R primer. If you use an older version of the scheme it could affect your analysis, 7 bases that could result from primers might be included in your final consensus so you should update your workflows.
It is advisable to update to the latest version of wf-artic when you start using the new kit version.
Folder structure
The growing number of supported schemes mean that we needed to reorganise the primer_schemes folder. The new structure is shown below. Briefly, under each scheme name of SARS-CoV-2 or spike-seq a folder exists that separates the schemes into the main assays, be that ARTIC, Midnight-ONT, or the newly added Midnight-IDT. Within each of these folders are the primer versions available for that assay. The scheme name therefore remains SARS-CoV-2, but the version now includes the assay and version of that assay, e.g Midnight-ONT/V3.
data/primer_schemes├── SARS-CoV-2│ ├── ARTIC│ │ ├── V1│ │ │ ├── SARS-CoV-2.reference.fasta│ │ │ └── SARS-CoV-2.scheme.bed│ │ ├── V2│ │ │ ├── SARS-CoV-2.reference.fasta│ │ │ └── SARS-CoV-2.scheme.bed│ │ ├── V3│ │ │ ├── SARS-CoV-2.reference.fasta│ │ │ └── SARS-CoV-2.scheme.bed│ │ ├── V4│ │ │ ├── SARS-CoV-2.reference.fasta│ │ │ └── SARS-CoV-2.scheme.bed│ │ └── V4.1│ │ ├── SARS-CoV-2.reference.fasta│ │ └── SARS-CoV-2.scheme.bed│ ├── Midnight-IDT│ │ └── V1│ │ ├── SARS-CoV-2.reference.fasta│ │ └── SARS-CoV-2.scheme.bed│ ├── Midnight-ONT│ │ ├── V1│ │ │ ├── SARS-CoV-2.reference.fasta│ │ │ └── SARS-CoV-2.scheme.bed│ │ ├── V2│ │ │ ├── SARS-CoV-2.reference.fasta│ │ │ └── SARS-CoV-2.scheme.bed│ │ └── V3│ │ ├── SARS-CoV-2.reference.fasta│ │ └── SARS-CoV-2.scheme.bed│ └── NEB-VarSkip��│ ├── v1a│ │ ├── SARS-CoV-2.reference.fasta│ │ └── SARS-CoV-2.scheme.bed│ ├── v1a-long│ │ ├── SARS-CoV-2.reference.fasta│ │ └── SARS-CoV-2.scheme.bed│ └── v2│ ├── SARS-CoV-2.reference.fasta│ └── SARS-CoV-2.scheme.bed└── spike-seq└── ONT├── V1│ ├── spike-seq.reference.fasta│ ├── spike-seq.scheme.bed│ └── spike-seq.vcf└── V4.1├── spike-seq.reference.fasta├── spike-seq.scheme.bed└── spike-seq.vcf
Running wf-artic
To facilitate the existence of potentially multiple primers schemes wf-artic allows for the specification of the scheme at run time.
nextflow run epi2me-labs/wf-artic \--fastq <PATH_TO_MIDNIGHT-ONT_V3_FASTQ> \--scheme_name SARS-CoV-2 \--scheme_dir primer_schemes \--scheme_version MIDNIGHT-ONT/V3
To list the schemes that are packaged with wf-artic you can run:
nextflow run epi2me-labs/wf-artic \--fastq <PATH_TO_FASTQ> \--list_schemes
Custom primer schemes
We understand that users may want to supply their own schemes for use with wf-artic analysis. To enable this we have added the command line argument --custom_scheme. This must be the full path to the directory containing your appropriately named scheme bed and fasta files; <SCHEME_NAME>.bed and <SCHEME_NAME>.fasta.
You must specify the scheme name (--scheme_name) and the min (--min_len) and max (--max_len) length parameters based on your expected amplicons for read length filtering.
You must provide both a fasta of your genome and a primer bed file detailing your primers, please use the ones included in wf-artic as examples; These must be readable and contained within the directory you specify for --custom-scheme. We do not validate your bed or fasta file so please ensure these are in the correct format.

