Targeted BRCA Gene Analysis with Oxford Nanopore
Oxford Nanopore Technologies products are not intended for use for health assessment or to diagnose, treat, mitigate, cure or prevent any disease or condition.
wf-human-variation is our “do it all” Nextflow workflow for the identification of human DNA variation from Oxford Nanopore Technologies’ long read sequencing data. With its latest update, ClinVar variant annotation is now also included.
This post will show that our human variation workflow can also process targeted sequence data. In the following text we will analyse targeted BRCA1 and BRCA2 sequence data that was generated in a research setting using Oxford Nanopore Technologies’ adaptive sampling methodology. The analysis will be performed with our EPI2ME software running on a laptop computer. While the BRCA gene enrichment here was prepared using adaptive sampling, this “how to” would also apply to capture and/or PCR based methods of enrichment. The analysis described here could similarly be applied to large panels of genes of interest in research environments.
Data was generated using DNA from the cell line NA14636 provided by the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research.
NA14636 sample is from a 56 year-old female with family history of breast cancer. This individual was diagnosed with the disease at 50. We know that certain mutations in BRCA1 and BRCA2 can result in increased risk of hereditary breast and ovarian cancer, and so we sequenced the sample to see if we could detect any key BRCA1 and BRCA2 mutations.
The sample was prepared and sequenced using our ligation sequencing kit and the newer Ligation Sequencing Kit V14 on a GridION for 72hrs, with adaptive sampling of BRCA1 and BRCA2.
Analysis
Let’s dive in and and see how we can get from our long Oxford Nanopore sequencing reads to a list of interesting variants.
If you haven’t already downloaded EPI2ME it’s freely available for all major operating systems here: https://labs.epi2me.io/downloads/, there are some pre-requisites but the application will take you through these.
Inputs
Our 1st task is to make sure we have the required input files for the workflow. The required inputs to wf-human-variation are the following:
- a BED file of regions we are interested in analysing (target regions)
- a BAM file of mapped or unmapped sequencing reads (MinKNOW, dorado generated (or other))
- a FASTA reference genome
BAM file(s)
If you don’t have a mapped/unmapped BAM file input please see the appendix for instructions.
BED file
If you have performed adaptive sampling you can simply use the BED file you used as input into MinKNOW when setting up your sequencing run, if not then you can easily make a BED file, it is a tab separated file with 3 mandatory columns of the chromosome, start and end position of your region. You can add additional columns for the name and size of the region for instance. More details on the BED format here
The BED file for the analysis described here looks like this:
chr13 32305479 32409671 BRCA2 104192chr17 43034294 43135363 BRCA1 101069
FASTA reference genome
Download the human reference genome - we’ll use hg38 from UCSC in this example:
https://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/analysisSet/hg38.analysisSet.fa.gz
Running wf-human-variation
We can easily analyse data from small panels locally on a reasonably powered laptop, or on a GridION or PromethION device using the EPI2ME interface. Our workflow for the analysis of human variation, wf-human-variation, is available in the application and is easy to install and run (Figure 1).
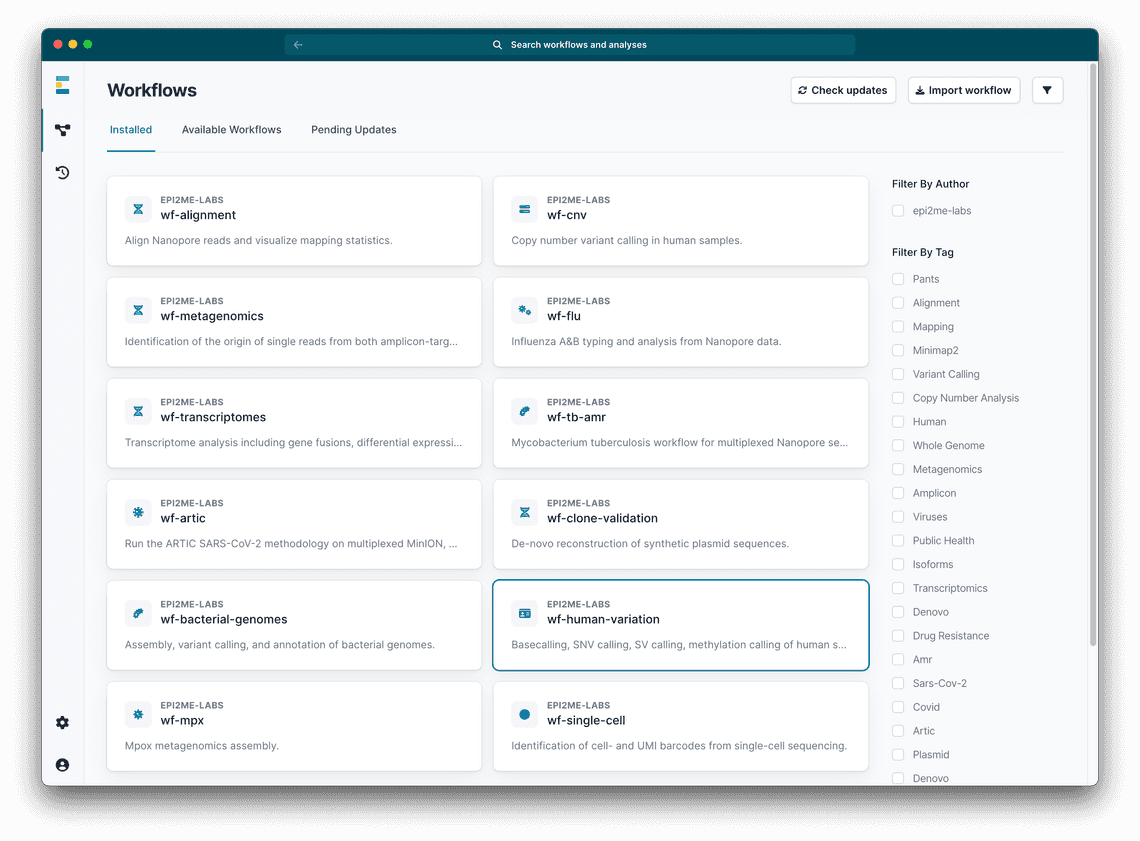
In “Workflow Options” choose “Snp” - annotation of SNPs is now carried out automatically (
>= v1.6.0).In “Input Options” choose:
- the
BAMfile you created withwf-alignmentor any pre-generated BAM file you wish to analyse. - the
FASTAreference genome - the
BEDfile defining the regions you wish to analyse
Under “Small variant calling options” choose “Phase VCF” - ONT long reads mean we can determine the haplotype of the variants called
Again you can alter the resources given to the workflow under “Multiprocessing Options” and “Extra Configuration”
Click “Launch workflow”
Results
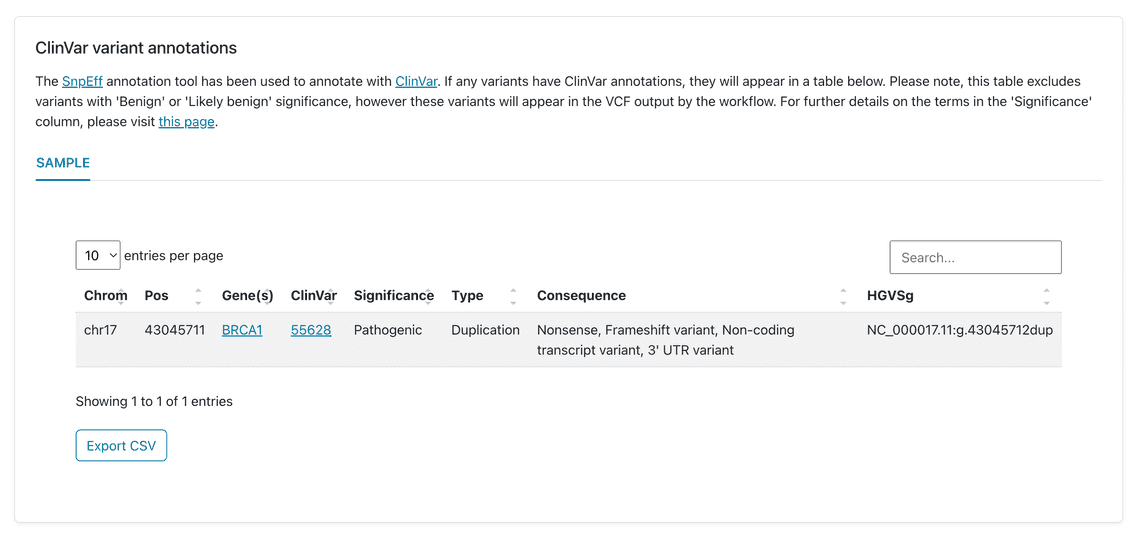
Using ClinVar to annotate the variants found by wf-human-variation (Figure 2) highlights one pathogenic variant in BRCA1 which is a 1bp insertion at nucleotide 5677 in exon 24 (5677insA). This results in a frameshift and truncation at codon 1853 (Y1853X). This matches the information provided by Coriell for this sample.
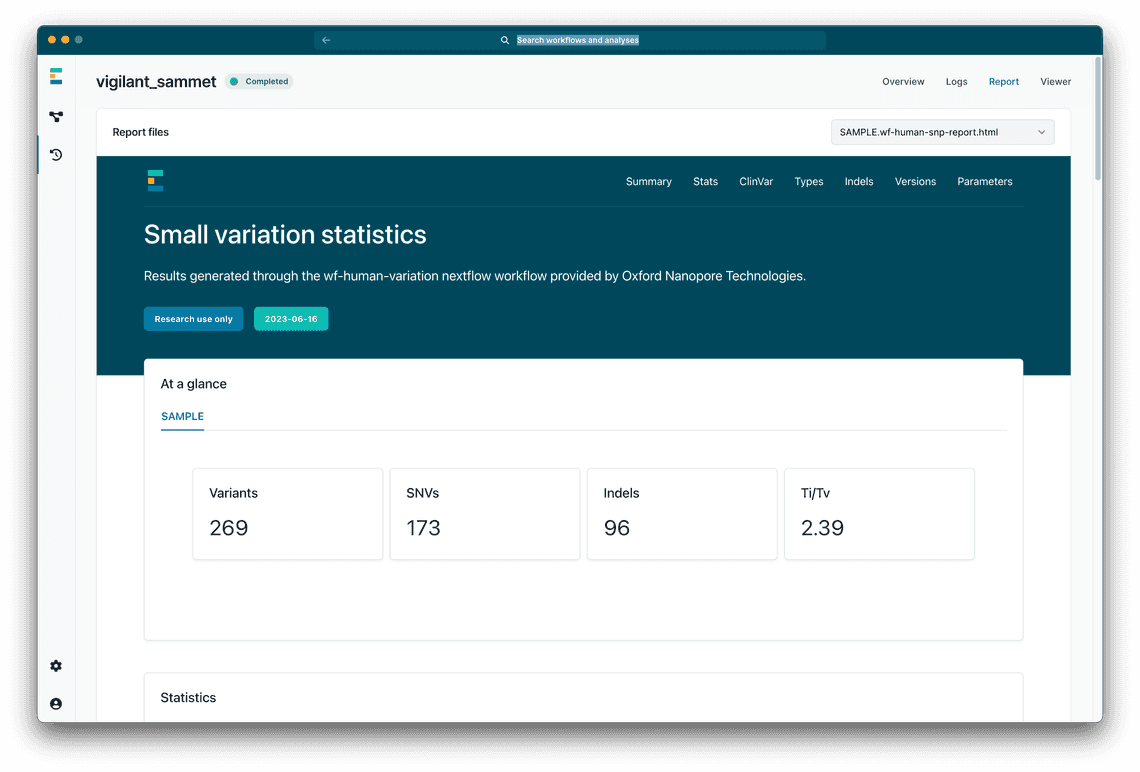
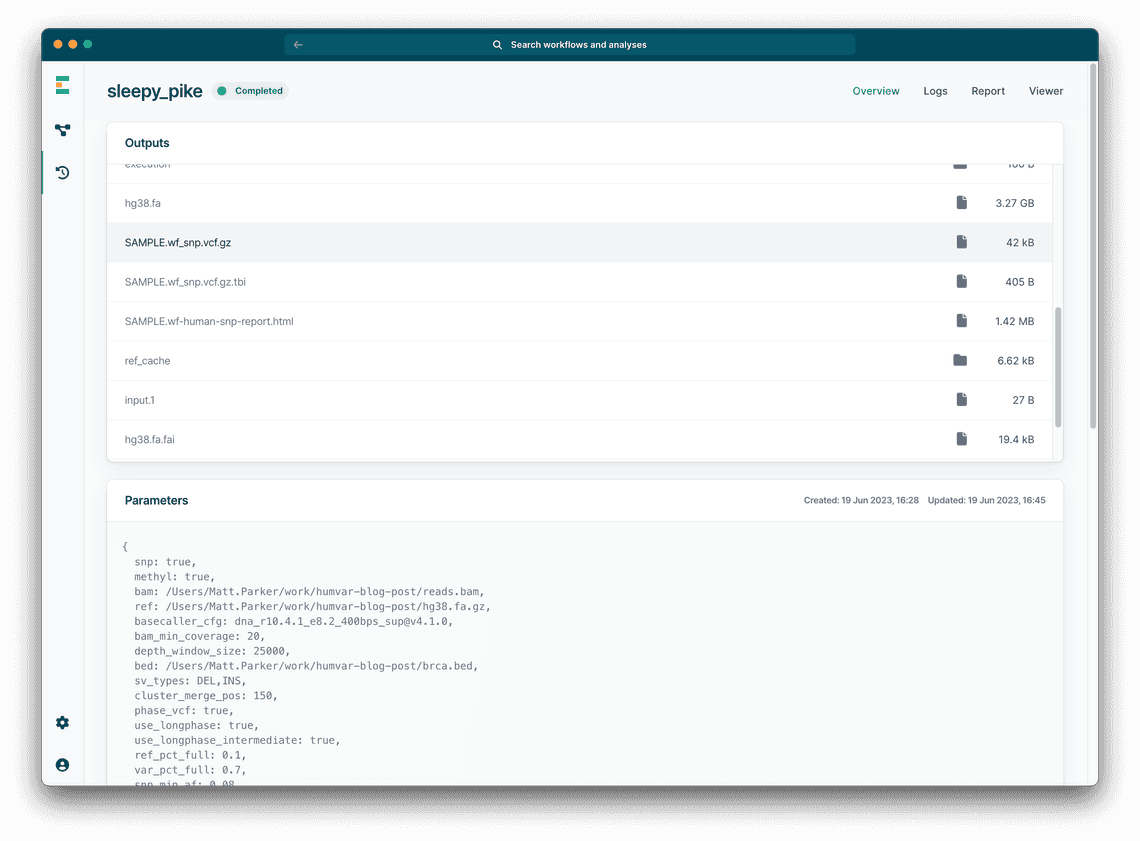
Other important data is output by the workflow (Figure 3), including a summaries for the variants, and the alignment in HTML format.
You can also take a look at the raw data such as the VCF file for the variants (Figure 4). Clicking “Open folder” at the bottom of the page will open a file explorer to see the raw data.
Conclusion
We hope that you found this quick tutorial on how to analyse targeted human Oxford Nanopore sequencing research data with wf-human-variation and EPI2ME. Any comments, questions or suggestions don’t hesitate to let us know using the usual channels.
Appendix 1 - BAM file generation
You have two options if you have FASTQ data:
- Align your reads
- Make an unmapped bam
1. Alignment with wf-alignment
You’ll need the reference genome FASTA from above; we can align our sequencing reads from our FASTQ files to this reference with wf-alignment to generate some statistics on our sequencing data and also create our BAM file for wf-human-variation.
In EPI2ME Labs install the wf-alignment workflow if you haven’t already done so. Click “Run this workflow”
Choose the path to your FASTQ files and the path to your reference genome; hg38.analysisSet.fa.gz under “Input Options”.
You can also increase the resources given to the workflow under “Misc Options” and “Extra Configuration”.
Click “Launch workflow”
2. Make an unmapped BAM
If you are reasonably familiar with the command line you can make what’s called an “unmapped” BAM file using samtools
samtools import -o umapped_reads.bam -O BAM <FASTQ>
Where <FASTQ> is the path to your FASTQ file(s). This command will produce an umapped BAM file called unmapped_reads.bam.
Appendix 2 - Running on the command line
Just like with our desktop application, running wf-human-variation on the command line is easy.
A good place to start is to list the parameters that can be used to run the workflow:
nextflow run epi2me-labs/wf-human-variation --help
To recreate the analysis above, these are the options to use:
nextflow run epi2me-labs/wf-human-variation --bam <PATH_TO_READS> --ref <PATH_TO_FASTA> --bed <PATH_TO_BED> --phase_vcf --snp
Table Of Contents
Related Posts
Information

