An update on our Copy Number Analysis workflow
We have previously shared our standalone workflow for performing copy number analysis. The standalone version has been deprecated, and the functionality from this workflow has been incorporated into wf-human-variation, so we would recommend users switch over to using this sub-workflow.
The main functionality of the sub-workflow remains the same, with QDNAseq at its core. QDNAseq is an R package which determines the copy number status of bins, the size of which can be tuned by using the --bin_size parameter at run time. Pre-calculated bin annotations are available for hg19 and hg38 for a range of bin sizes (1, 5, 10, 15, 30, 50, 100, 500, and 1000 kbp). If --bin_size is not specified then a default of 500 is used. QDNAseq, is based on the commonly-used read depth strategy, which correlates the copy number of a region with the depth of coverage, so for example, a gain in copy number would have a higher depth than expected.
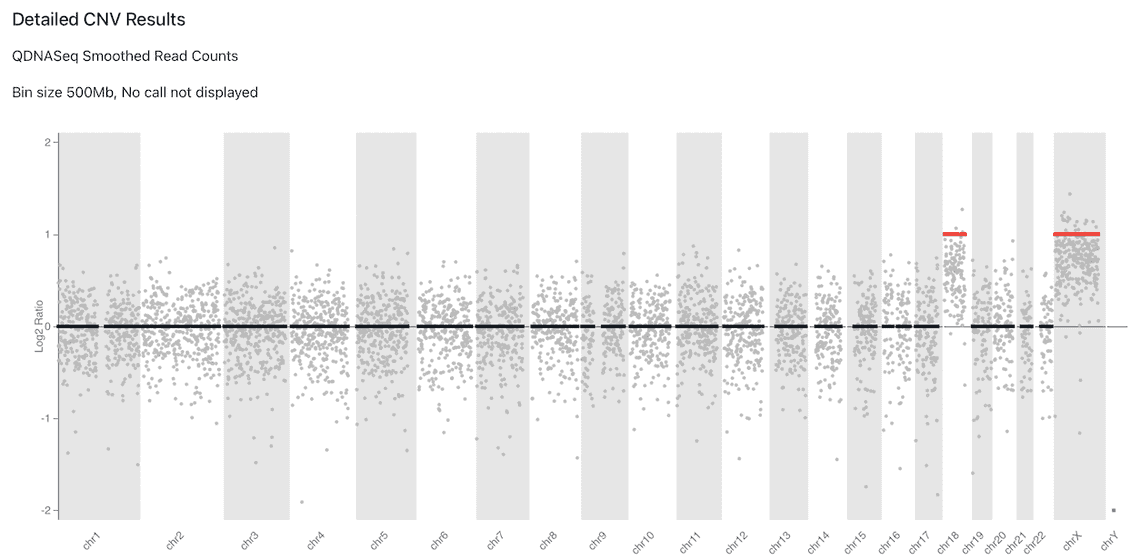
The sub-workflow outputs an HTML report, and Figure 1 shows an example of a copy number ideoplot from the report generated by running this sub-workflow. This example has resulted from the analysis of NA03623, a cell line sample obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research characterised as trisomy X and trisomy 18.
Running the workflow
As CNV calling is now part of wf-human-variation, the example command has been updated accordingly:
nextflow run epi2me-labs/wf-human-variation --cnv --bam <PATH_TO_BAM> --ref <PATH_TO_REFERENCE> --bin_size <BIN_SIZE>
A note on bin size selection
If the chosen bin size is incorrect, you may see the following R error when running the workflow:
Calculating correction for GC content and mappability2 Error in getGlobalsAndPackages(expr, envir = envir, globals = globals) :3 The total size of the 26 globals exported for future expression ('FUN()') is 778.60 MiB.. This exceeds the maximum allowed size of 500.00 MiB (option 'future.globals.maxSize'). The three largest globals are 'object' (435.98 MiB of class 'S4'), 'counts' (282.55 MiB of class 'numeric') and 'gc' (23.56 MiB of class 'numeric')4 Calls: estimateCorrection ... getGlobalsAndPackagesXApply -> getGlobalsAndPackages5 Execution halted
To assist with resolving this, the Applications team have provided some recommended bin sizes based on a 3.2Gb genome, which we are pleased to share below:
| Bin size | Minimum read count (20/bin) | Optimal read count (200/bin) |
|---|---|---|
| 15 | 4266666 | 42666666 |
| 30 | 2133333 | 21333333 |
| 50 | 1280000 | 12800000 |
| 100 | 640000 | 6400000 |
| 500 | 128000 | 1280000 |
| 1000 | 64000 | 640000 |
If the R error above is encountered, then please adjust the --bin_size parameter accordingly. Recommendations for bin size may evolve in the future, and we will endeavour to keep the community up to date with best practice.
Reference
- Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, van Essen HF, Eijk PP, Rustenburg F, Meijer GA, Reijneveld JC, Wesseling P, Pinkel D, Albertson DG, Ylstra B. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014 Dec;24(12):2022-32. doi: 10.1101/gr.175141.114. Epub 2014 Sep 18. PMCID.
Share
Sirisha Hesketh
Clinical Bioinformatician
Table Of Contents
Related Posts
Information
