How to align your data

Read alignment is central to most bioinformatics analyses. In this short post we explain how to align reads against a reference and analyse the resulting files.
Read alignment isn’t usually an end point of analysis; its typically combined with subsequent steps to obtain useful information. Readers might be interested in some of our workflows that include read alignment as an integral step:
- wf-alignment - for a simple alignment of your reads with one or more references.
- wf-amplicon - for alignment of amplicon sequence data with relevant references.
- wf-transcriptomes - for alignment and subsequent analysis of cDNA and direct RNA data.
All will output a sorted aligned BAM file as well as other useful statistics and an easy to interpret report. Our preferred tool for alignment is Minimap2.
Minimap2
We advise using Minimap2 for alignment of long reads to a reference. There are various publications (eg. LoTempio et al., 2021, Becht et al., 2021) that compare alignment tools using real and simulated reads for a variety of applications. Minimap2 is typically faster than other tools whilst maintaining general good accuracy across different types of Oxford Nanopore Technologies’ read data.
Minimap2 is a fast sequence mapping and alignment program. It works by first mapping reads to the approximate origin of the reference sequence, and then running a computationally intensive comparison between localised reads to create the final alignments. You will often see these two steps referred to together as alignment. If you are interested in further detail on how minimap2 works you might like to read the author’s paper.
Using minimap2 with nanopore data
Let’s break down the basic invocation of the tool for genomic reads:
minimap2 -a -x map-ont ref.fa query.fq > alignment.sam
- minimap2 - is the base command.
- -a - a flag to turn on SAM output, otherwise minimap2 outputs a PAF format by default.
- -x map-ont - use the map-ont preset will set parameters with suitable defaults.
- ref.fa - refers to the reference: the sequence or sequences against which your reads will be aligned. This can also be a precomputed
.mmiindex file. - query.fq - is the FASTQ file containing your reads.
- > alignment.sam - the output will be directed to this SAM file.
You should not adjust any parameters as they have been fine tuned with tested defaults using the -x map-ont preset.
If working with cDNA use the -x splice preset instead eg.
minimap2 -a -x splice ref.fa query-cDNA.fq > alignment-cDNA.sam
How to post process the alignment
A few common steps might be required to get the alignments in a useful form.
The first of these is to convert the SAM file to a more space efficient BAM (-S for SAM, -b for BAM):
samtools view -S -b alignment.sam > alignment.bam
We can also sort the alignments by reference position to create a sorted BAM:
samtools sort -o alignment.sorted.bam alignment.bam
And we can index the sorted alignments file to allow quick retrieval of reads spanning a given reference position:
samtools index alignment.sorted.bam
It is possible to combine all of these steps in a single incantation using pipes:
minimap2 -a -x map-ont ref.fa query.fq \samtools sort -O BAM --write-index -o alignment.sorted.bam -
How to analyse alignments.
Minimap2 can output three types of alignment for long reads, they are:
- Primary alignments - the alignment with the highest alignment score. There should only ever be one primary alignment per read.
- Secondary alignments - a read may align ambiguously to multiple locations, any additional possible alignments will be secondary alignments.
- Supplementary alignments - the aligner may break a read into two or more pieces and aligns both, perhaps due to a structural variation. The best aligned segment of the chain will be the primary alignment while the others will be marked as supplementary. Supplementary alignments are also of key importance when analyses cDNA or direct RNA reads aligned to a genomic reference (or incomplete transcriptome).
- Unmapped - if the read cannot be aligned it is still included in the alignment file and marked as unmapped.
A good way to get a quick summary report of the types of alignment records in your aligned bam file is with samtools flagstat
samtools flagstat sorted.alignment.bam12000 + 0 in total (QC-passed reads + QC-failed reads)5000 + 0 secondary3000 + 0 supplementary0 + 0 duplicates10000 + 0 mapped (93.98% : N/A)0 + 0 paired in sequencing0 + 0 read10 + 0 read20 + 0 properly paired (N/A : N/A)0 + 0 with itself and mate mapped0 + 0 singletons (N/A : N/A)0 + 0 with mate mapped to a different chr0 + 0 with mate mapped to a different chr (mapQ>=5)
This shows that there are 10000 primary alignments indicated by mapped and 2000 unmapped (total - mapped = unmapped). Other categories may or may not be relevant depending on your use case --- many of those above relate specifically to paired-end short-read sequencing.
More detailed per-read information can be viewed using samtools view
samtools view alignment.bam
The SAM/BAM specification provides a detailed explanation of the information available in the alignment file but some main points are the name of the read that was mapped, the position with respect to the reference, the sequence of the read and per base quality scores of the read.
One column which deserves additional comment (and people frequently ask us about), is the “mapping quality” (MAPQ) column. For minimap2 this is given as a score in the range 0-60: it is a phred-scaled probability of the alignment being incorrect. For example if the score is 60 it means we can consider the read uniquely mapped to a single region of the reference sequence. A lower values indicates that the read shows similarity to multiple regions of the reference sequence and cannot be uniquely assigned to a single region.
Samtools has many more tools to help filter, visualise and manipulate alignment files. Read more about samtools and the options available here. We also have a more detailed tutorial of working with SAM/BAM files available here.
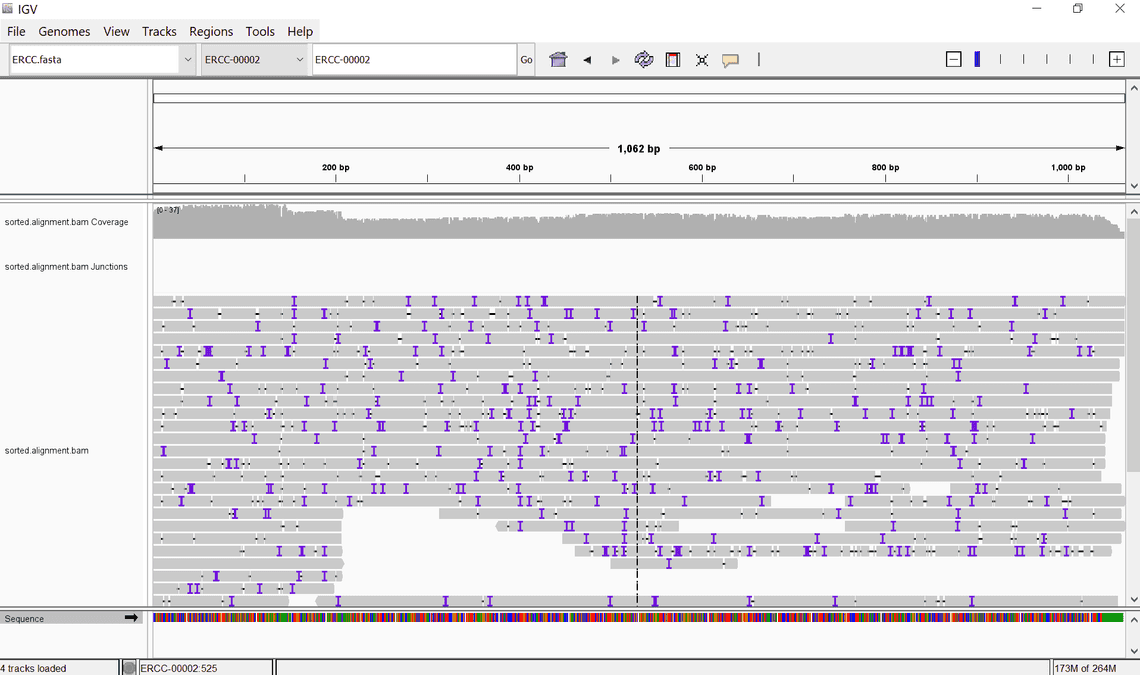
Another great way to make sense of your alignment files is to visualise them in an alignment viewer such as IGV or JBrowse. The minimum you will require for this is the fasta file you originally used for the alignment and the sorted, and indexed bam files we produced earlier with samtools.
Summary
Alignment is a crucial first step in many bioinformatic tasks. Here we have given a brief survey of the tools of the trade for creating and summarising alignments with nanopore data.
Table Of Contents
Related Posts
Information

