Supporting Future Clinical Bioinformaticians

Oxford Nanopore Technologies products are not intended for use for health assessment or to diagnose, treat, mitigate, cure or prevent any disease or condition.
Over the past couple of months the EPI2ME team have welcomed two trainee clinical bioinformaticians from the National Health Service in England (NHS) to spend some time working on small projects in the team.
Sophia Johnson and Benjamin Bunce are individuals on the NHS scientist training programme (STP), training to become clinical scientists specialising in bioinformatics. There are clinical scientists in numerous healthcare specialisms working behind the scenes in hospitals, and those on the Bioinformatics-Genomics pathway in the NHS are primarily responsible for the analysis of next-generation sequencing data. Based in regional laboratories, they work closely with scientists in specialist disease teams, to provide clinicians with diagnostic reports for their patients.
Matt Parker and I have previously worked in this role, providing safe and effective analysis of NGS data for the benefit of patient care.
We’ve loved having Sophia and Ben as guests in the team and we wish them every success in the remainder of their training. Sophia writes about her experience with us below.
Sophia Johnson - Repeat Expansion Interruptions
Spending 6 weeks at ONT with the EPI2ME team for my elective has been a great opportunity to broaden my Bioinformatics skills before starting my third and final year of the NHS STP.
The Project
My main project has been to create a new plot for the wf-human-variation STR report. Short Tandem Repeat (STR) expansions can cause diseases such as Huntington’s and Fragile X Syndrome. The plot I aimed to create needed to give a more complete picture of an individual’s STRs through displaying base content of each read and highlighting any interruptions.
wf-human-variation utilises a fork of Straglr to genotype STR expansions from long-read alignments. The Staglr tool outputs a VCF and a TSV file with detailed information per read. The BAM file and Straglr TSV from the workflow would provide all the information required for me to create the plot.
Weeks 1-2
I spent my first two weeks getting up to speed with the EPI2ME codebase. I also worked through tutorials around Nextflow and various plotting tools that could be of use such as Apache ECharts and Bokeh. Personally, I find tutorials and Youtube videos are often good starting points before reading the tools documentation in more detail.
- Youtube: Nextflow Training Workshop Playlist
- Youtube: Demo of various Apache ECharts
- Tutorial: Introduction to NextFlow
- Tutorial: NextFlow Basic Concepts
Weeks 3-4
My next focus was the data pre-processing steps required for the plot. Here�’s an overview of the main steps…
- Filtering the workflow BAM:
- For speed purposes I first used samtools to subset the BAM to contain only STR regions
- Next I filtered the BAM to contain only STR supporting reads from the Straglr TSV (utilising Pandas and pysam)
- Extracting STR sequences from the reads:
- First, relevant information was extracted from the Straglr TSV such as haplotype, repeat start position, and repeat size
- Supporting read sequences were extracted from the BAM
- STR sequences were then extracted from the read sequences
- Next I created lists of indexes to identify repeat units and interruptions within the STR sequences (utilising Regex)
- All the information was then added to a summary TSV that could be used as input for the plot
Week 5
I spent week 5 working on the code for the plot itself. We decided to utilise Bokeh as it provided the best starting point for the plot. Progress pictures below…
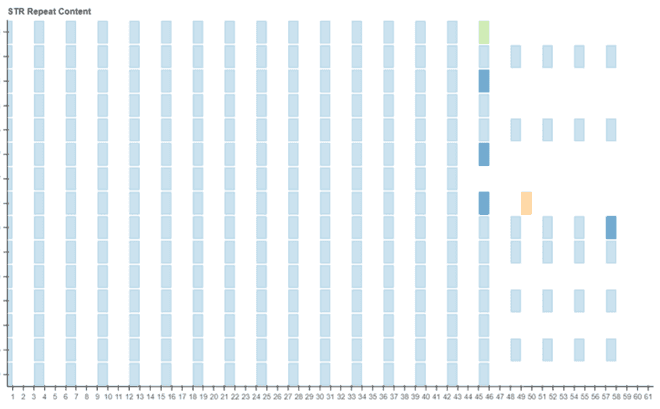
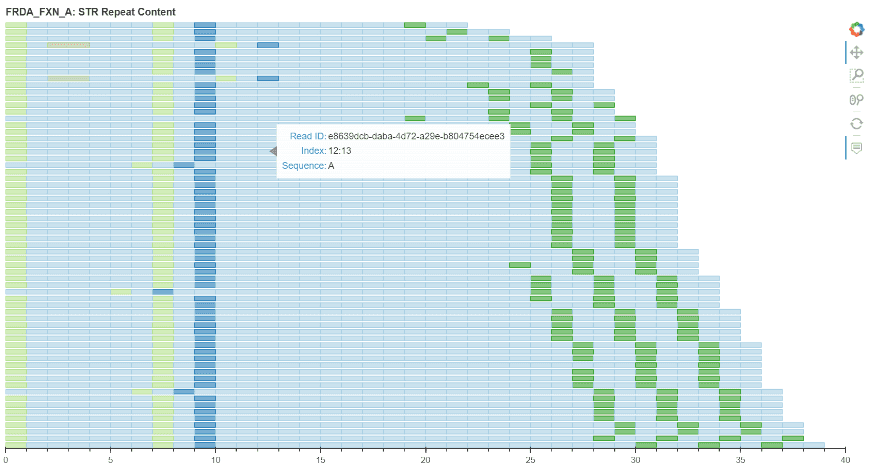
Week 6
Finally, in week 6 I worked on integrating my code into EPI2ME’s wf-human-variation. This involved converting my Python code into Nextflow and thinking about how best to display the plot to the users.
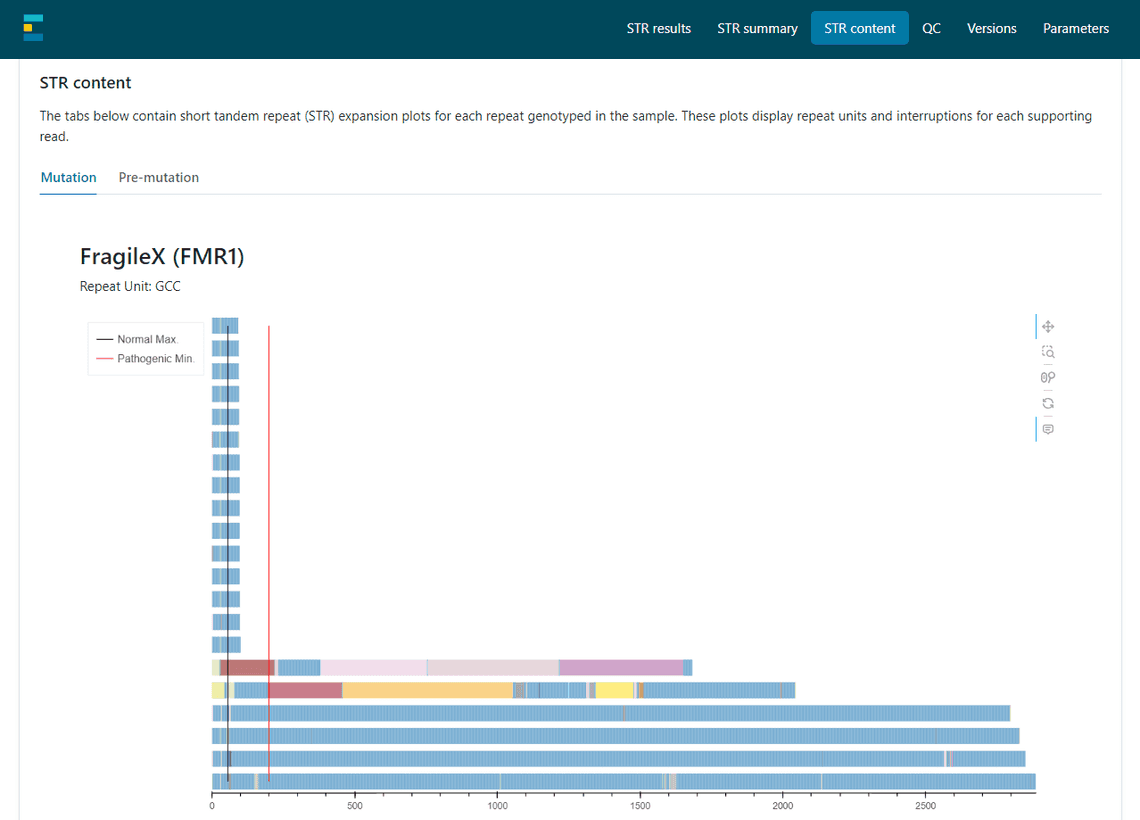
Other Highlights
Outside of gaining new technical skills through the repeat expansion project it has also been really interesting to spend time with a team of Industry Bioinformaticians. Industry and Clinical Bioinformaticians have slightly different considerations in their day-to-day work, however, many of the technical and project management tools that we utilise overlap, and so there are always things we can learn from each other. As the workflows developed by the EPI2ME team are open-source and utilised by many people outside their department (not just internally as in the NHS) they have a strong focus on user feedback and usability testing. We also have different accreditation standards that we need to meet.
Another highlight of the elective was having the opportunity to visit the ONT labs during my first week. One of ONT’s Technical Product Managers showed me how to undertake library preparation and load a GridION Flow Cell. It was fascinating to see the real-time sequencing analysis via MinKNOW (Watching things like the channel states panel and the cumulative output). The next day I was able to access the sequencing data which I could then run through an EPI2ME workflow to see the whole end-to-end process.
Overall, this has been a really valuable experience and I want to thank the EPI2ME team for having me!

