Short Tandem Repeat expansion genotyping in wf-human-variation
Our human variation workflow, wf-human-variation, gives users the ability to call a range of different variant types from Oxford Nanopore Technologies sequencing data, offering: SNP, SV, methylation, and CNV sub-workflows. In this blog post we introduce the newest sub-workflow, which enables genotyping of STR expansions.
Our colleagues in the ONT Applications Team are actively investigating best practice for human STR genotyping, and this sub-workflow builds on their extensive work to make this functionality available to the community.
STR expansion genotyping
Short tandem repeats (STRs) are short DNA sequence motifs, typically 2-6bp, which are repeated consecutively at given positions in the genome. An abnormal number of copies of these motifs at certain loci have been shown to cause human disease, for example trinucleotide repeat expansion disorders such as Huntington’s disease (HTT) and Fragile X syndrome (FMR1). As long-read ONT sequencing data has the potential to cover the whole expanded region, it is particularly well-suited to both identification of expansions as well as calculating their size - a critical aspect of determining repeat instability and a notable advantage over short-read sequencing platforms.
Running the workflow
The STR workflow implemented within wf-human-variation accepts BAM as input along with a (optionally gzipped) FASTA reference sequence, and uses a fork of Straglr to genotype STRs and generate a VCF, followed by annotation via Stranger and SnpSift. Genotyping is based on a BED file of repeats, available here.
Example command
Please note, this workflow is only compatible with human genome build 38.
nextflow run epi2me-labs/wf-human-variation --str --bam <PATH_TO_BAM> --ref <PATH_TO_REFERENCE> --sex <male|female>
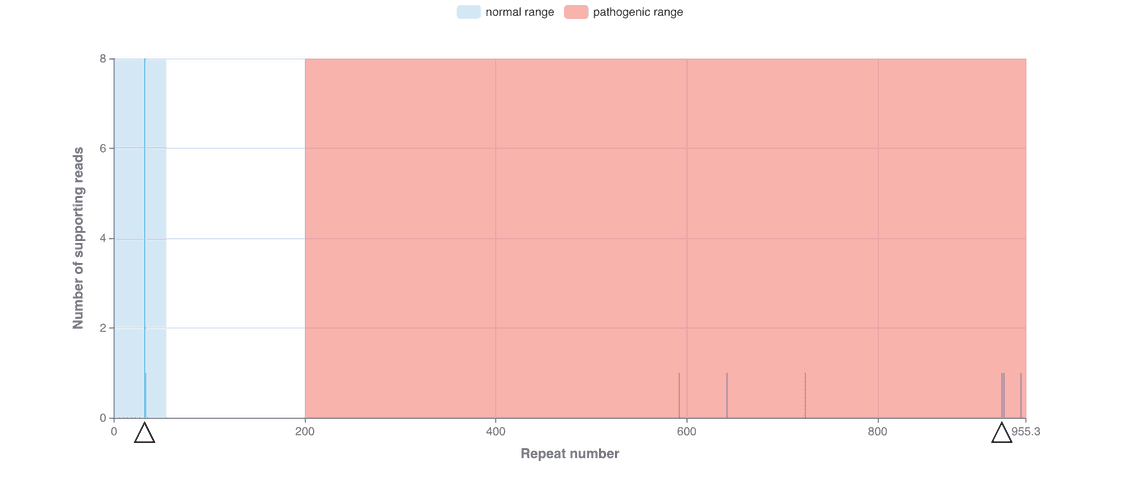
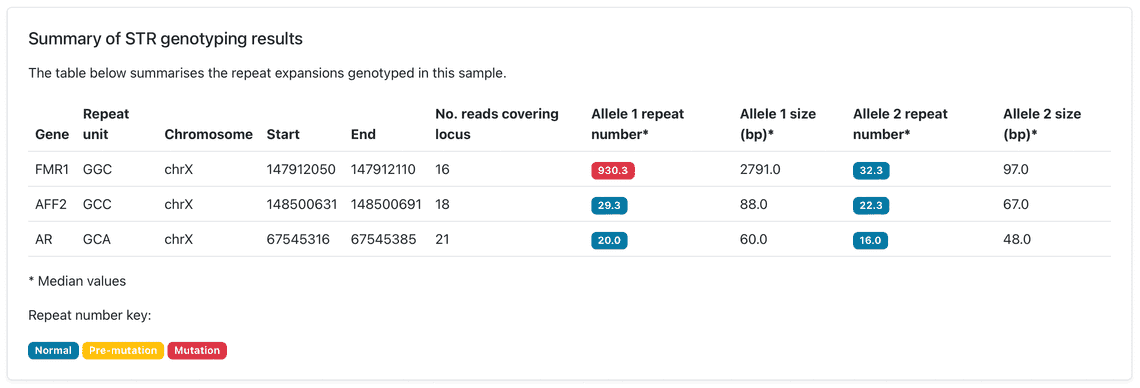
The STR workflow triggers a modified version of the SNP workflow to create a haplotagged BAM, as phased alignments are required to genotype STR expansions, enabling the enumeration of repeat units and their corresponding lengths for each allele. A haplotagged BAM is only generated if the user selects the --str option, and won’t be generated if --snp is selected on its own. The STR workflow also takes a required --sex parameter (male or female) which determines the number of calls on chrX. The output HTML report contains a list of each repeat, divided into ‘normal’, ‘pre-mutation’ and ‘pathogenic’ tabs, depending on the number of copies of the repeat observed in each haplotype (Figure 1). A summary table is also presented which gives an at-a-glance overview of the total number of each repeat, again colour-coded according to whether the repeat number falls into the normal, pre-mutation or pathogenic ranges (Figure 2). These examples have been observed following an analysis of sequence data from the Coriell NA07063 cell line.
As well as the HTML report, the workflow also produces as output the final haplotagged BAM used for STR genotyping, an annotated VCF of calls, and a tab-delimited file output by Straglr which lists the reads spanning STR sites, and includes read identifiers and strand information.
References
Chiu, R., Rajan-Babu, IS., Friedman, J.M. et al. Straglr: discovering and genotyping tandem repeat expansions using whole genome long-read sequences. Genome Biol 22, 224 (2021). https://doi.org/10.1186/s13059-021-02447-3
The following cell line samples were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research: NA07063
Share
Sirisha Hesketh
Clinical Bioinformatician
Table Of Contents
Related Posts
Information
