Nextflow in Windows with WSL2
For EPI2ME Labs version 3 and onward the setup of Windows Subsytem for Linux is automated provided users have an up-to-date version of Windows 10 or 11. These instructions are provided to aid users on older versions of Windows that cannot update. For users who can update we (and Microsoft) recommend doing so before attempting to use the method presented here. The simple wsl --install method should be used in most cases, which is the method used in the automated setup within EPI2ME Labs 3 and newer. The methods here are not officially supported.
It is easy to run and develop Nextflow workflows on Windows but there are some additional set up steps required. We have found the set up can be tricky so have outlined the steps required here with links to trouble shooting and further help.
This guide is similar to that presented at a guide from Nextflow which goes in to greater detail and has additional steps useful for developing workflows.
You will need Windows 10 and it is critical that you have Admin privileges for all the steps,
Check your windows version is sufficient
- Start > search ‘Windows update settings’ > at the bottom select ‘OS Build info’
- Under Windows specifications - Under OS Build info - Your Build number must be 18362.1049 or 18363.1049 (or higher).
- Make any Windows updates that are required, this will cause your computer to restart and could take a while.
Enable Windows Subsystem for Linux (WSL).
If you have Windows 10 May 2020 (2004) update (or later) installed you can install WSL with a single command:
- Start > search for ‘powershell’ > Right click and run as admin > copy/paste the following -
wsl.exe --install- Restart your computer and you will be prompted to set up Ubuntu with a username and password and can skip to step 8.
If you have an earlier Windows version or the above has not worked continue following the steps -
- Start > search for ‘powershell’ > Right click and run as admin > copy/paste the following -
dism.exe /online /enable-feature /featurename:Microsoft-Windows-Subsystem-Linux /all /norestart- Copy and paste the following in to Powershell
dism.exe /online /enable-feature /featurename:VirtualMachinePlatform /all /norestartUpdate to WSL2
Download this WSL2 update file and run through the installation wizard: wsl_update.msi
A guide to WSL installation troubleshooting can be found here.
Set WSL2 as the default version.
- Open a new Powershell with admin rights
- In Powershell copy and paste
wsl --set-default-version 2Download WSL Ubuntu
- From the start menu open the Microsoft store
- In search bar type Ubuntu
- Download and install the Ubuntu 64-bit version: Focal 20.04 (LTS).
Check WSL version for Ubuntu installation
WSL installations come in two versions, creatively name 1 and 2. We must ensure the Ubuntu installation is set to version 2.
In Powershell run:
wsl --list –vThe response should look something like:
H:\> wsl --list -vNAME STATE VERSION* Ubuntu Running 2docker-desktop-data Running 2docker-desktop Running 2Ubuntu-18.04 Running 1If a version is listed as 1 and not 2, change the version with
wsl --set-version <distribution name> <version>For example to change the Ubuntu-18.04 installation above to version 2:
H:\> wsl --set-version Ubuntu 2
Set up Docker
- Download Docker Desktop
- Run the installation as an administrator and follow the install wizard.
- Add your computer username to the “docker group” in WSL - in Powershell type:
wslsudo groupadd dockersudo usermod -aG docker $(whoami)- Reboot your PC
Enable WSL2 in docker
These docker instructions can also be found here
- Start Docker Desktop from the Windows Start menu.
- From the Docker menu, select Settings > General
- The ‘Use the WSL 2 based engine’ box should already be ticked, if not tick the box and click Apply & Restart.
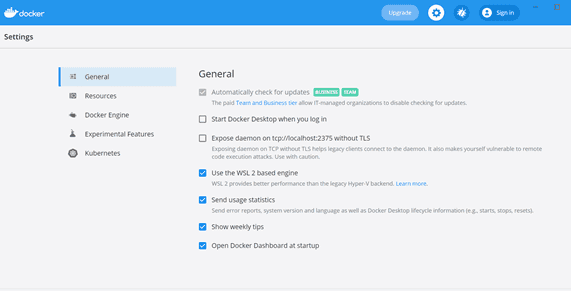
Docker settings - Go to Settings > Resources > WSL Integration
- ‘Enable integration with my default WSL distro’ should be ticked and select Enable integration with the Ubuntu version you have just downloaded.
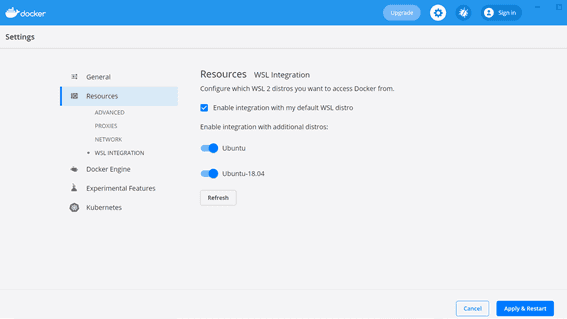
Docker WSL Integration - If you are unsure of the Ubuntu version go to Powershell and type
wsllsb_release -dThis will respond with something like:
Description: Ubuntu 20.04.3 LT0Set WSL2 resource limits
It is important to configure docker resource limits so it does not use all available memory and cpu.
You will need to update a different file depending on if you want a global configurartion for all WSL distributions or a specific one for each distribution.
Global
- In Windows Explorer navigate to
C:/Users/<USERNAME> - Create or open existing file
.wslconfig
- In Windows Explorer navigate to
Specific Distribution
- Open specific distribution terminal eg. Ubuntu or type wsl Powershell
- Navigate to the
/etcand open a file:
cd /etcsudo vim wsl.confCopy and paste the following in to the respective file
[wsl2]memory=8GBprocessors=4localhostForwarding=true
Adjust settings in the config based on your available resources. To find out total available on your device you can CTRL-ALT-DELETE to open task manager, click performance tab and look at details of CPU and memory. Total no. of processors will be ‘logical processors’. Remember to leave some resource available for other programmes on your computer.
Save the file (in vim press Esc and type
:wq!).Shut down all running wsl instances via cmd line
wsl --shutdownNow relaunch wsl and check resources are as expected using cmds
cat /proc/cpuinfocat /proc/meminfo
For more advanced settings see here
Install Java
To install Java into the WSL2 Ubuntu installation run (after starting WSL with the
wslcommand):sudo apt install openjdk-11-jre-headlessDownload and install Nextflow
Nextflow can be installed following the instruction in their Getting started pages:
- Run the following:
curl -s https://get.nextflow.io | bashmv nextflow /usr/bin/nextflowchmod +x /usr/bin/nextflow- Make sure Nextflow is updated to the latest version by typing -
nextflow self-updateTest nextflow
To test Nextflow we can run a simple workflow template.
- To obtain some test data, download the workflow:
git clone https://github.com/epi2me-labs/wf-template.git- To see required parameters of the workflow run:
nextflow run epi2me-labs/wf-template- And to run the workflow with the test data run:
OUTPUT=outputnextflow run epi2me-labs/wf-template \-w ${OUTPUT}/workspace \--fastq wf-template/test_data --out_dir wf-template/${OUTPUT}
If you had any problems with this tutorial or think any additional information would be useful then please let us know so we can improve it for other users.

